Gene
KWMTBOMO11354 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001966
Annotation
aldehyde_dehydrogenase_isoform_2_[Bombyx_mori]
Full name
Aldehyde dehydrogenase
Location in the cell
Cytoplasmic Reliability : 1.875
Sequence
CDS
ATGACAGTCGGGAATCTTACAACATCCAAACAAAAGACGATGTCAGCTGCTGAGGCCGTCCAAAAAGCTCGAGACACTTTCAATCGCGGCACAACCAGGCCGATAGAATGGCGCCGTCAGCAGCTCAAGAATTTGTTAAGAATGTACGAGGAAAACCAAAACGTCATGGTGGAGGCCCTACACAAGGATCTCAGACGAAGCAAAATGGAAGCCATTCTACTCGAAGTCGACTACTTGATCAATGATTTGAGGAACACTCTGCATTATTTGGATGAGTGGACTAAGCCGGAACATCCTCCAAAAGGCTTCGTGAACATTCTAGACGAGGTGGTTATATACAATGACCCGTACGGCGTCGTCCTCGTAATAGGCGCCTGGAACTATCCCTTGCAGCTGCTGCTTTTGCCGATGGCCGGAGCCATCGCTGCCGGGAACACGGTTATCGTGAAACCCAGCGAACTATCAGTCGCCTGCTCGAACTTTGTTGTCGAAAATCTACCGAAATATTTGGATAATGATGCTTTCATCGTATTCGAAGGAGGTCCGCAAGAGACCACTGAGTTGCTCAAACAGAGGTTTGACTACATCTTCTACACTGGAGGCACTAACGTCGGTAGAATTGTTTACGAAGCCGCCACCAAAAATCTCACGCCTGTCACATTGGAACTTGGCGGCAAAAGCCCAGTTTACGTAGATAACACGGTGGACATAGTTGTGACGGCAAAACGAATTCTCTGGGGTAAATTTATAAACGTGGGCCAAACTTGCATTGCTCCCGACTACGTACTGTGCACGAAGGAGGTACAGAACAAGTTTCTGGAGGCCTCCAAGAAAGTCCTGAAGGAATGGTACGGAGAGGACCCACAGAAATCCCCCGATCTATGCCGTATCATAAATAACAGGCACTTCAGCCGTCTGCAGCAGCTGATAGAATCGAGCAAGAACAAAATAGCCATCGGTGGTTGTTACGACTCTAACGACAGATATATAGAGCCCACAATATTGGCCAACGTCACGCCGGCCGACAAAGTGATGGAAGACGAGATATTCGGTCCCATCTTACCAATAGTTTGTATTGAAAACGCTTACGAGGCCATCAAATTTATAAATGAAAGGGAACATCCGCTAGTGCTTTACGTGTTCAGCAATCAATACAACATACAAAAGTTATTCACTGAACAGACTCGCTCTGGAAGCATTAGTATCAACGACACGATCATGTTTTACGGCGTTGAAACTCTACCGTTTGGTGGTGTGGGTAACAGCGGTATAGGTGCGTACCACGGAAAGAAGACTATCGATACATTCACACACAAAAAGAGTTGCCTCAAGAAGAACTTCGCTGCTATTGGAGAGAAACTGGCTTCAGGCCGCTACCCTCCTTACACAGAAGGAAACTTGAAATTCATAACGGCACTAATGAAGAAACGTCACGGGCCCTCGCTAAAGTATCTGCCACACTTCATGGCCTTCGCTCTCGGAGCTGCTATATCTTACGGTATCTTCGCATGGCAAAAGATGGCGTCCGATGATTCTTAA
Protein
MTVGNLTTSKQKTMSAAEAVQKARDTFNRGTTRPIEWRRQQLKNLLRMYEENQNVMVEALHKDLRRSKMEAILLEVDYLINDLRNTLHYLDEWTKPEHPPKGFVNILDEVVIYNDPYGVVLVIGAWNYPLQLLLLPMAGAIAAGNTVIVKPSELSVACSNFVVENLPKYLDNDAFIVFEGGPQETTELLKQRFDYIFYTGGTNVGRIVYEAATKNLTPVTLELGGKSPVYVDNTVDIVVTAKRILWGKFINVGQTCIAPDYVLCTKEVQNKFLEASKKVLKEWYGEDPQKSPDLCRIINNRHFSRLQQLIESSKNKIAIGGCYDSNDRYIEPTILANVTPADKVMEDEIFGPILPIVCIENAYEAIKFINEREHPLVLYVFSNQYNIQKLFTEQTRSGSISINDTIMFYGVETLPFGGVGNSGIGAYHGKKTIDTFTHKKSCLKKNFAAIGEKLASGRYPPYTEGNLKFITALMKKRHGPSLKYLPHFMAFALGAAISYGIFAWQKMASDDS
Summary
Similarity
Belongs to the aldehyde dehydrogenase family.
Belongs to the NDK family.
Belongs to the NDK family.
Uniprot
Q2F5I9
Q2F5J0
H9IXI3
A0A2W1BQ95
A0A3S2LZM4
A0A2H1W1Q4
+ More
A0A194RB61 A0A1L8D714 A0A194QVM6 A0A212F520 A0A1Q3FLE4 A0A1Q3FL55 A0A1Q3FKX3 M9T154 B0W5G6 A0A2A4JQY4 A0A1Q3FL16 M9T6D2 M9SYR6 A0A1Q3FKY1 A0A1Q3FL24 A0A2J7RN02 A0A2J7RMZ6 A0A2J7RN13 K7J3Y9 A0A2J7RMZ1 M9T4M3 Q16MV5 A0A023EUE7 A0A182N808 A0A182UNC1 A0A1S4GNY9 A0A182H762 A0A1S4FVG5 M9T0L1 Q7PQ09 A0A182ISI2 A0A182LBE0 U5EUY4 A0A182JTN8 A0A232F9B4 A0A182WAA8 A0A1L8DZT0 A0A084W9K4 A0A1Q3FL79 A0A1L8E0A1 A0A453YRU6 A7UT82 A0A1L8E005 A0A1Q3FLE5 A0A182LSR1 A0A453YRN7 A0A182RTP2 A0A2M3Z8G5 S4PB99 A0A2M4BL12 A0A2M4BKP9 A7UT81 W5J9E3 D2SNX0 A0A2M4AKZ2 A0A453YJT1 A0A2M4CNA2 A0A2M4CNL1 A0A453YJI2 W8BT84 A0A0A1WJM9 W8B5M1 A0A1B6CUY8 A0A0C9RAF2 A0A182PHD7 A0A1B6CPG8 A0A1I8PAD2 A0A1B6E8W0 A0A1W4V0G7 A0A158NEB0 A0A1B6CRG9 A0A0A1WVN3 A0A1W4VCR3 A0A195CCD8 A0A151J6X4 A0A1I8MCL6 A0A0A1X9V8 A0A1W4VE28 A0A0B4KEE1 E3CTS3 A0A1J1I8P8 Q7JR61 A0A034VVW2 A0A034VXI2 A4UZ69 A0A0C4FEI5 A0A034VXI8 T1PLC3 A0A034VVW7 T1PCX5 Q8IGH3 A0A0B4KF99 A0A0B4KEL0 A1Z6Z4
A0A194RB61 A0A1L8D714 A0A194QVM6 A0A212F520 A0A1Q3FLE4 A0A1Q3FL55 A0A1Q3FKX3 M9T154 B0W5G6 A0A2A4JQY4 A0A1Q3FL16 M9T6D2 M9SYR6 A0A1Q3FKY1 A0A1Q3FL24 A0A2J7RN02 A0A2J7RMZ6 A0A2J7RN13 K7J3Y9 A0A2J7RMZ1 M9T4M3 Q16MV5 A0A023EUE7 A0A182N808 A0A182UNC1 A0A1S4GNY9 A0A182H762 A0A1S4FVG5 M9T0L1 Q7PQ09 A0A182ISI2 A0A182LBE0 U5EUY4 A0A182JTN8 A0A232F9B4 A0A182WAA8 A0A1L8DZT0 A0A084W9K4 A0A1Q3FL79 A0A1L8E0A1 A0A453YRU6 A7UT82 A0A1L8E005 A0A1Q3FLE5 A0A182LSR1 A0A453YRN7 A0A182RTP2 A0A2M3Z8G5 S4PB99 A0A2M4BL12 A0A2M4BKP9 A7UT81 W5J9E3 D2SNX0 A0A2M4AKZ2 A0A453YJT1 A0A2M4CNA2 A0A2M4CNL1 A0A453YJI2 W8BT84 A0A0A1WJM9 W8B5M1 A0A1B6CUY8 A0A0C9RAF2 A0A182PHD7 A0A1B6CPG8 A0A1I8PAD2 A0A1B6E8W0 A0A1W4V0G7 A0A158NEB0 A0A1B6CRG9 A0A0A1WVN3 A0A1W4VCR3 A0A195CCD8 A0A151J6X4 A0A1I8MCL6 A0A0A1X9V8 A0A1W4VE28 A0A0B4KEE1 E3CTS3 A0A1J1I8P8 Q7JR61 A0A034VVW2 A0A034VXI2 A4UZ69 A0A0C4FEI5 A0A034VXI8 T1PLC3 A0A034VVW7 T1PCX5 Q8IGH3 A0A0B4KF99 A0A0B4KEL0 A1Z6Z4
Pubmed
19121390
28756777
26354079
22118469
23639754
20075255
+ More
17510324 24945155 12364791 26483478 14747013 17210077 20966253 28648823 24438588 23622113 20920257 23761445 20074338 24495485 25830018 21347285 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373
17510324 24945155 12364791 26483478 14747013 17210077 20966253 28648823 24438588 23622113 20920257 23761445 20074338 24495485 25830018 21347285 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373
EMBL
DQ311434
ABD36378.1
DQ311433
ABD36377.1
BABH01018195
BABH01018196
+ More
BABH01018197 BABH01018198 KZ149982 PZC75794.1 RSAL01000096 RVE47726.1 ODYU01005766 SOQ46953.1 KQ460436 KPJ14729.1 GEYN01000005 JAV02124.1 KQ461073 KPJ09588.1 AGBW02010285 OWR48809.1 GFDL01006739 JAV28306.1 GFDL01006822 JAV28223.1 GFDL01006784 JAV28261.1 KC243495 AGI96738.1 DS231842 EDS35286.1 NWSH01000823 PCG74014.1 GFDL01006797 JAV28248.1 KC243497 AGI96740.1 KC243496 AGI96739.1 GFDL01006808 JAV28237.1 GFDL01006833 JAV28212.1 NEVH01002546 PNF42206.1 PNF42205.1 PNF42209.1 AAZX01001008 PNF42204.1 KC243498 AGI96741.1 CH477847 EAT35690.1 GAPW01001052 JAC12546.1 AAAB01008900 JXUM01115632 JXUM01115633 JXUM01115634 JXUM01115635 JXUM01115636 JXUM01115637 KQ565901 KXJ70553.1 KC243499 AGI96742.1 EAA09458.5 GANO01001252 JAB58619.1 NNAY01000677 OXU27049.1 GFDF01002091 JAV11993.1 ATLV01021829 KE525324 KFB46898.1 GFDL01006718 JAV28327.1 GFDF01002092 JAV11992.1 EDO64087.1 GFDF01002097 JAV11987.1 GFDL01006711 JAV28334.1 AXCM01000363 GGFM01004040 MBW24791.1 GAIX01004711 JAA87849.1 GGFJ01004543 MBW53684.1 GGFJ01004505 MBW53646.1 EDO64086.1 ADMH02002078 ETN59470.1 EZ407255 ACX53811.1 GGFK01008096 MBW41417.1 APCN01002922 GGFL01002628 MBW66806.1 GGFL01002739 MBW66917.1 GAMC01014141 JAB92414.1 GBXI01015674 JAC98617.1 GAMC01014142 JAB92413.1 GEDC01019998 JAS17300.1 GBYB01005250 JAG75017.1 GEDC01022024 JAS15274.1 GEDC01002929 JAS34369.1 ADTU01013229 ADTU01013230 ADTU01013231 ADTU01013232 GEDC01021307 JAS15991.1 GBXI01011400 JAD02892.1 KQ978023 KYM97896.1 KQ979851 KYN18822.1 GBXI01006777 JAD07515.1 AE013599 AGB93305.1 BT125739 ADP21878.1 CVRI01000041 CRK95326.1 BT003248 BT003252 AAF59248.2 AAO25009.1 GAKP01012368 GAKP01012359 JAC46584.1 GAKP01012367 GAKP01012365 JAC46585.1 AAM68900.3 BT126071 AAM68894.5 ADY17770.1 GAKP01012370 GAKP01012362 GAKP01012361 JAC46590.1 KA649572 AFP64201.1 GAKP01012371 GAKP01012364 GAKP01012363 JAC46589.1 KA646611 AFP61240.1 BT001784 AAN71539.1 AGB93308.1 AGB93306.1 AAM68899.3
BABH01018197 BABH01018198 KZ149982 PZC75794.1 RSAL01000096 RVE47726.1 ODYU01005766 SOQ46953.1 KQ460436 KPJ14729.1 GEYN01000005 JAV02124.1 KQ461073 KPJ09588.1 AGBW02010285 OWR48809.1 GFDL01006739 JAV28306.1 GFDL01006822 JAV28223.1 GFDL01006784 JAV28261.1 KC243495 AGI96738.1 DS231842 EDS35286.1 NWSH01000823 PCG74014.1 GFDL01006797 JAV28248.1 KC243497 AGI96740.1 KC243496 AGI96739.1 GFDL01006808 JAV28237.1 GFDL01006833 JAV28212.1 NEVH01002546 PNF42206.1 PNF42205.1 PNF42209.1 AAZX01001008 PNF42204.1 KC243498 AGI96741.1 CH477847 EAT35690.1 GAPW01001052 JAC12546.1 AAAB01008900 JXUM01115632 JXUM01115633 JXUM01115634 JXUM01115635 JXUM01115636 JXUM01115637 KQ565901 KXJ70553.1 KC243499 AGI96742.1 EAA09458.5 GANO01001252 JAB58619.1 NNAY01000677 OXU27049.1 GFDF01002091 JAV11993.1 ATLV01021829 KE525324 KFB46898.1 GFDL01006718 JAV28327.1 GFDF01002092 JAV11992.1 EDO64087.1 GFDF01002097 JAV11987.1 GFDL01006711 JAV28334.1 AXCM01000363 GGFM01004040 MBW24791.1 GAIX01004711 JAA87849.1 GGFJ01004543 MBW53684.1 GGFJ01004505 MBW53646.1 EDO64086.1 ADMH02002078 ETN59470.1 EZ407255 ACX53811.1 GGFK01008096 MBW41417.1 APCN01002922 GGFL01002628 MBW66806.1 GGFL01002739 MBW66917.1 GAMC01014141 JAB92414.1 GBXI01015674 JAC98617.1 GAMC01014142 JAB92413.1 GEDC01019998 JAS17300.1 GBYB01005250 JAG75017.1 GEDC01022024 JAS15274.1 GEDC01002929 JAS34369.1 ADTU01013229 ADTU01013230 ADTU01013231 ADTU01013232 GEDC01021307 JAS15991.1 GBXI01011400 JAD02892.1 KQ978023 KYM97896.1 KQ979851 KYN18822.1 GBXI01006777 JAD07515.1 AE013599 AGB93305.1 BT125739 ADP21878.1 CVRI01000041 CRK95326.1 BT003248 BT003252 AAF59248.2 AAO25009.1 GAKP01012368 GAKP01012359 JAC46584.1 GAKP01012367 GAKP01012365 JAC46585.1 AAM68900.3 BT126071 AAM68894.5 ADY17770.1 GAKP01012370 GAKP01012362 GAKP01012361 JAC46590.1 KA649572 AFP64201.1 GAKP01012371 GAKP01012364 GAKP01012363 JAC46589.1 KA646611 AFP61240.1 BT001784 AAN71539.1 AGB93308.1 AGB93306.1 AAM68899.3
Proteomes
UP000005204
UP000283053
UP000053240
UP000007151
UP000002320
UP000218220
+ More
UP000235965 UP000002358 UP000008820 UP000075884 UP000075903 UP000069940 UP000249989 UP000007062 UP000075880 UP000075882 UP000075881 UP000215335 UP000075920 UP000030765 UP000075883 UP000075900 UP000000673 UP000075840 UP000075885 UP000095300 UP000192221 UP000005205 UP000078542 UP000078492 UP000095301 UP000000803 UP000183832
UP000235965 UP000002358 UP000008820 UP000075884 UP000075903 UP000069940 UP000249989 UP000007062 UP000075880 UP000075882 UP000075881 UP000215335 UP000075920 UP000030765 UP000075883 UP000075900 UP000000673 UP000075840 UP000075885 UP000095300 UP000192221 UP000005205 UP000078542 UP000078492 UP000095301 UP000000803 UP000183832
Pfam
Interpro
IPR016161
Ald_DH/histidinol_DH
+ More
IPR012394 Aldehyde_DH_NAD(P)
IPR016163 Ald_DH_C
IPR016162 Ald_DH_N
IPR029510 Ald_DH_CS_GLU
IPR015590 Aldehyde_DH_dom
IPR001806 Small_GTPase
IPR027417 P-loop_NTPase
IPR003578 Small_GTPase_Rho
IPR005225 Small_GTP-bd_dom
IPR016160 Ald_DH_CS_CYS
IPR001012 UBX_dom
IPR029071 Ubiquitin-like_domsf
IPR036241 NSFL1C_SEP_dom_sf
IPR012989 SEP_domain
IPR023005 Nucleoside_diP_kinase_AS
IPR036850 NDK-like_dom_sf
IPR034907 NDK-like_dom
IPR001564 Nucleoside_diP_kinase
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR025136 DUF4071
IPR017441 Protein_kinase_ATP_BS
IPR000719 Prot_kinase_dom
IPR012394 Aldehyde_DH_NAD(P)
IPR016163 Ald_DH_C
IPR016162 Ald_DH_N
IPR029510 Ald_DH_CS_GLU
IPR015590 Aldehyde_DH_dom
IPR001806 Small_GTPase
IPR027417 P-loop_NTPase
IPR003578 Small_GTPase_Rho
IPR005225 Small_GTP-bd_dom
IPR016160 Ald_DH_CS_CYS
IPR001012 UBX_dom
IPR029071 Ubiquitin-like_domsf
IPR036241 NSFL1C_SEP_dom_sf
IPR012989 SEP_domain
IPR023005 Nucleoside_diP_kinase_AS
IPR036850 NDK-like_dom_sf
IPR034907 NDK-like_dom
IPR001564 Nucleoside_diP_kinase
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR025136 DUF4071
IPR017441 Protein_kinase_ATP_BS
IPR000719 Prot_kinase_dom
SUPFAM
ProteinModelPortal
Q2F5I9
Q2F5J0
H9IXI3
A0A2W1BQ95
A0A3S2LZM4
A0A2H1W1Q4
+ More
A0A194RB61 A0A1L8D714 A0A194QVM6 A0A212F520 A0A1Q3FLE4 A0A1Q3FL55 A0A1Q3FKX3 M9T154 B0W5G6 A0A2A4JQY4 A0A1Q3FL16 M9T6D2 M9SYR6 A0A1Q3FKY1 A0A1Q3FL24 A0A2J7RN02 A0A2J7RMZ6 A0A2J7RN13 K7J3Y9 A0A2J7RMZ1 M9T4M3 Q16MV5 A0A023EUE7 A0A182N808 A0A182UNC1 A0A1S4GNY9 A0A182H762 A0A1S4FVG5 M9T0L1 Q7PQ09 A0A182ISI2 A0A182LBE0 U5EUY4 A0A182JTN8 A0A232F9B4 A0A182WAA8 A0A1L8DZT0 A0A084W9K4 A0A1Q3FL79 A0A1L8E0A1 A0A453YRU6 A7UT82 A0A1L8E005 A0A1Q3FLE5 A0A182LSR1 A0A453YRN7 A0A182RTP2 A0A2M3Z8G5 S4PB99 A0A2M4BL12 A0A2M4BKP9 A7UT81 W5J9E3 D2SNX0 A0A2M4AKZ2 A0A453YJT1 A0A2M4CNA2 A0A2M4CNL1 A0A453YJI2 W8BT84 A0A0A1WJM9 W8B5M1 A0A1B6CUY8 A0A0C9RAF2 A0A182PHD7 A0A1B6CPG8 A0A1I8PAD2 A0A1B6E8W0 A0A1W4V0G7 A0A158NEB0 A0A1B6CRG9 A0A0A1WVN3 A0A1W4VCR3 A0A195CCD8 A0A151J6X4 A0A1I8MCL6 A0A0A1X9V8 A0A1W4VE28 A0A0B4KEE1 E3CTS3 A0A1J1I8P8 Q7JR61 A0A034VVW2 A0A034VXI2 A4UZ69 A0A0C4FEI5 A0A034VXI8 T1PLC3 A0A034VVW7 T1PCX5 Q8IGH3 A0A0B4KF99 A0A0B4KEL0 A1Z6Z4
A0A194RB61 A0A1L8D714 A0A194QVM6 A0A212F520 A0A1Q3FLE4 A0A1Q3FL55 A0A1Q3FKX3 M9T154 B0W5G6 A0A2A4JQY4 A0A1Q3FL16 M9T6D2 M9SYR6 A0A1Q3FKY1 A0A1Q3FL24 A0A2J7RN02 A0A2J7RMZ6 A0A2J7RN13 K7J3Y9 A0A2J7RMZ1 M9T4M3 Q16MV5 A0A023EUE7 A0A182N808 A0A182UNC1 A0A1S4GNY9 A0A182H762 A0A1S4FVG5 M9T0L1 Q7PQ09 A0A182ISI2 A0A182LBE0 U5EUY4 A0A182JTN8 A0A232F9B4 A0A182WAA8 A0A1L8DZT0 A0A084W9K4 A0A1Q3FL79 A0A1L8E0A1 A0A453YRU6 A7UT82 A0A1L8E005 A0A1Q3FLE5 A0A182LSR1 A0A453YRN7 A0A182RTP2 A0A2M3Z8G5 S4PB99 A0A2M4BL12 A0A2M4BKP9 A7UT81 W5J9E3 D2SNX0 A0A2M4AKZ2 A0A453YJT1 A0A2M4CNA2 A0A2M4CNL1 A0A453YJI2 W8BT84 A0A0A1WJM9 W8B5M1 A0A1B6CUY8 A0A0C9RAF2 A0A182PHD7 A0A1B6CPG8 A0A1I8PAD2 A0A1B6E8W0 A0A1W4V0G7 A0A158NEB0 A0A1B6CRG9 A0A0A1WVN3 A0A1W4VCR3 A0A195CCD8 A0A151J6X4 A0A1I8MCL6 A0A0A1X9V8 A0A1W4VE28 A0A0B4KEE1 E3CTS3 A0A1J1I8P8 Q7JR61 A0A034VVW2 A0A034VXI2 A4UZ69 A0A0C4FEI5 A0A034VXI8 T1PLC3 A0A034VVW7 T1PCX5 Q8IGH3 A0A0B4KF99 A0A0B4KEL0 A1Z6Z4
PDB
1AD3
E-value=7.36504e-134,
Score=1223
Ontologies
PATHWAY
00010
Glycolysis / Gluconeogenesis - Bombyx mori (domestic silkworm)
00053 Ascorbate and aldarate metabolism - Bombyx mori (domestic silkworm)
00071 Fatty acid degradation - Bombyx mori (domestic silkworm)
00280 Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
00310 Lysine degradation - Bombyx mori (domestic silkworm)
00330 Arginine and proline metabolism - Bombyx mori (domestic silkworm)
00340 Histidine metabolism - Bombyx mori (domestic silkworm)
00380 Tryptophan metabolism - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00561 Glycerolipid metabolism - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
00981 Insect hormone biosynthesis - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00053 Ascorbate and aldarate metabolism - Bombyx mori (domestic silkworm)
00071 Fatty acid degradation - Bombyx mori (domestic silkworm)
00280 Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
00310 Lysine degradation - Bombyx mori (domestic silkworm)
00330 Arginine and proline metabolism - Bombyx mori (domestic silkworm)
00340 Histidine metabolism - Bombyx mori (domestic silkworm)
00380 Tryptophan metabolism - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00561 Glycerolipid metabolism - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
00981 Insect hormone biosynthesis - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
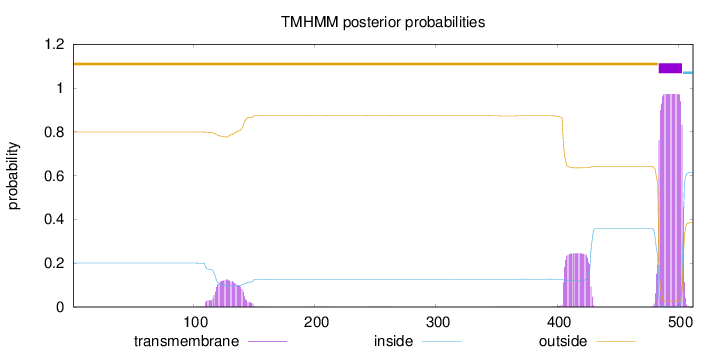
Topology
Length:
512
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
28.4855
Exp number, first 60 AAs:
0
Total prob of N-in:
0.20096
outside
1 - 483
TMhelix
484 - 503
inside
504 - 512
Population Genetic Test Statistics
Pi
239.719481
Theta
159.681371
Tajima's D
1.156803
CLR
0.405759
CSRT
0.705564721763912
Interpretation
Uncertain
