Gene
KWMTBOMO11351 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001853
Annotation
ATP_synthase_[Bombyx_mori]
Full name
ATP synthase subunit alpha
+ More
ATP synthase subunit alpha, mitochondrial
ATP synthase subunit alpha, mitochondrial
Alternative Name
ATP synthase F1 sector subunit alpha
F-ATPase subunit alpha
Protein bellwether
F-ATPase subunit alpha
Protein bellwether
Location in the cell
Mitochondrial Reliability : 3.804
Sequence
CDS
ATGTCGCTGATCTCCGCTCGTATTGCTGGCTCTGTAGCCAGGCGCTTGCCTAATGCTGCTACCCAGGTGTCAAAGGTGGCGGTGCCCGCAGTGGCCGTGGCATCTCGCAAACTACATGTCTCAACCACCCACAAAGCTGCCGAGATCTCCACCATCCTCGAAGAGAGGATCCTTGGAGCCGCGCCCAAGGCTGATCTAGAAGAGACTGGTCGTGTCTTGAGCATTGGTGATGGTATCGCTCGTGTTTATGGCTTGAAGAACATCCAGGCTGAGGAGATGGTGGAGTTCTCCTCAGGCCTCAAGGGAATGGCCCTTAACTTGGAACCTGACAATGTGGGTGTGGTAGTATTTGGTAATGACAAGCTTATCAAGGAAGGAGATATTGTCAAGCGTACTGGTGCTATCGTAGACGTTCCCGTCGGAGAGCAGATCCTTGGGCGTGTAGTAGATGCTTTGGGTAACCCTATTGATGGCAAGGGACCAATCGACACGAAATCCCGTATGAGGGTCGGTATTAAGGCGCCAGGTATCATTCCCCGGGTGTCTGTGCGTGAGCCTATGCAGACTGGTATCAAGGCTGTTGACTCTCTGGTACCAATTGGCCGTGGTCAGCGTGAGTTGATCATTGGTGACCGTCAAACTGGTAAGACTGCCCTGGCCATTGATACGATCATCAACCAGCAGAGATTCAACAAGGGTGAGGATGAGAAGAAGAAGTTGTACTGCATTTATGTTGCCATTGGACAGAAGAGGTCTACTGTAGCTCAAATTGTGAAGAGATTGACTGATGCTGGTGCCATCAACTACACAATCATCGTGTCTGCCACCGCTTCCGATGCTGCTCCCCTGCAATACCTGGCTCCTTACTCTGGTTGTGCCATGGGTGAATTTTTCCGTGATAATGGCAAACACGCCCTTATTATCTACGACGATTTGTCCAAACAAGCCGTAGCCTACCGTCAGATGTCTCTACTGCTGCGTCGTCCCCCCGGCCGTGAGGCTTATCCTGGTGACGTGTTCTATCTTCACTCCCGTCTGCTTGAGCGTGCCGCTAAGATGTCTGACAAAATGGGTGGTGGCTCCCTGACTGCTTTACCTGTCATTGAAACCCAAGCCGGTGATGTGTCAGCTTACATTCCTACCAACGTGATCTCCATCACTGATGGACAGATCTTCTTGGAAACTGAGCTGTTCTACAAAGGTATCCGACCAGCCATCAACGTCGGTCTGTCTGTATCGCGTGTCGGATCTGCTGCTCAGACCAAGGCTATGAAGCAGGTGGCCGGTTCCATGAAGCTGGAATTGGCTCAGTACCGTGAGGTCGCAGCTTTTGCCCAGTTCGGTTCTGACTTGGATGCCGCTACACAGCAGCTGCTCAACAGAGGAATGCGTCTTACTGAGCTCCTCAAGCAAGGACAATATGTGCCCATGGCTATTGAGGAACAGGTCGCCATCATTTACTGCGGTGTCCGCGGTCACCTCGACAAGCTGGACCCCTCCAAAATCACTGCCTTCGAGAAGGAATTCACTCAACACATCAAAACTAGCCACCAGGGTCTTCTCTCCACGATCGCCAAAGACGGTCAGATCACCCCCGAGTCTGACGCCTCCTTGAAGAAGATCGTCACAGACTTCCTAGCTACTTTCACCCAATCGCAGTAA
Protein
MSLISARIAGSVARRLPNAATQVSKVAVPAVAVASRKLHVSTTHKAAEISTILEERILGAAPKADLEETGRVLSIGDGIARVYGLKNIQAEEMVEFSSGLKGMALNLEPDNVGVVVFGNDKLIKEGDIVKRTGAIVDVPVGEQILGRVVDALGNPIDGKGPIDTKSRMRVGIKAPGIIPRVSVREPMQTGIKAVDSLVPIGRGQRELIIGDRQTGKTALAIDTIINQQRFNKGEDEKKKLYCIYVAIGQKRSTVAQIVKRLTDAGAINYTIIVSATASDAAPLQYLAPYSGCAMGEFFRDNGKHALIIYDDLSKQAVAYRQMSLLLRRPPGREAYPGDVFYLHSRLLERAAKMSDKMGGGSLTALPVIETQAGDVSAYIPTNVISITDGQIFLETELFYKGIRPAINVGLSVSRVGSAAQTKAMKQVAGSMKLELAQYREVAAFAQFGSDLDAATQQLLNRGMRLTELLKQGQYVPMAIEEQVAIIYCGVRGHLDKLDPSKITAFEKEFTQHIKTSHQGLLSTIAKDGQITPESDASLKKIVTDFLATFTQSQ
Summary
Description
Produces ATP from ADP in the presence of a proton gradient across the membrane.
Produces ATP from ADP in the presence of a proton gradient across the membrane. The alpha chain is a regulatory subunit.
Mitochondrial membrane ATP synthase (F(1)F(0) ATP synthase or Complex V) produces ATP from ADP in the presence of a proton gradient across the membrane which is generated by electron transport complexes of the respiratory chain. F-type ATPases consist of two structural domains, F(1) - containing the extramembraneous catalytic core, and F(0) - containing the membrane proton channel, linked together by a central stalk and a peripheral stalk. During catalysis, ATP synthesis in the catalytic domain of F(1) is coupled via a rotary mechanism of the central stalk subunits to proton translocation. Subunits alpha and beta form the catalytic core in F(1). Rotation of the central stalk against the surrounding alpha(3)beta(3) subunits leads to hydrolysis of ATP in three separate catalytic sites on the beta subunits. Subunit alpha does not bear the catalytic high-affinity ATP-binding sites (By similarity).
Produces ATP from ADP in the presence of a proton gradient across the membrane. The alpha chain is a regulatory subunit.
Mitochondrial membrane ATP synthase (F(1)F(0) ATP synthase or Complex V) produces ATP from ADP in the presence of a proton gradient across the membrane which is generated by electron transport complexes of the respiratory chain. F-type ATPases consist of two structural domains, F(1) - containing the extramembraneous catalytic core, and F(0) - containing the membrane proton channel, linked together by a central stalk and a peripheral stalk. During catalysis, ATP synthesis in the catalytic domain of F(1) is coupled via a rotary mechanism of the central stalk subunits to proton translocation. Subunits alpha and beta form the catalytic core in F(1). Rotation of the central stalk against the surrounding alpha(3)beta(3) subunits leads to hydrolysis of ATP in three separate catalytic sites on the beta subunits. Subunit alpha does not bear the catalytic high-affinity ATP-binding sites (By similarity).
Catalytic Activity
ATP + 4 H(+)(in) + H2O = ADP + 5 H(+)(out) + phosphate
Subunit
F-type ATPases have 2 components, CF(1) - the catalytic core - and CF(0) - the membrane proton channel. CF(1) has five subunits: alpha(3), beta(3), gamma(1), delta(1), epsilon(1). CF(0) has three main subunits: a, b and c.
Similarity
Belongs to the ATPase alpha/beta chains family.
Keywords
ATP synthesis
ATP-binding
Complete proteome
Direct protein sequencing
Hydrogen ion transport
Ion transport
Membrane
Mitochondrion
Mitochondrion inner membrane
Nucleotide-binding
Reference proteome
Transit peptide
Transport
Feature
chain ATP synthase subunit alpha, mitochondrial
Uniprot
Q2F5T3
A0A2H1W1X7
S4P2L2
A0A3S2LJ09
A0A194QXA3
A0A194QEQ8
+ More
A0A212ELG1 A0A2W1BNE3 A0A2A4JQE0 D6WSI9 A0A2J7Q0B3 A0A034WCI4 A0A0A1WN96 A0A0K8TW13 A0A067QI29 S4VDE6 W8CCY0 A0A182N7Z7 Q7PHI8 A0A182XJA6 A0A182UN55 A0A182LBD1 A0A182HP56 A0A182MRL5 A0A182WAB9 A0A191UR49 A0A084W9J4 Q1HRQ7 A0A182JM63 A0A0V0G2J4 A0A182PHE7 A0A1L8E154 A0A1J1J483 A0A182Q0H6 A0A182RTQ3 A0A0L0CN23 A0A069DZ92 B4MQA2 A0A182K4A3 A0A336N1U6 A0A323TPU0 A0A423TIL1 J3JWJ5 A0A0J9RIM8 A0A1A9XAQ8 A0A1B0BN74 B4I8B0 A0A1W4VDL2 A0A0N7ZCJ7 T1DEX4 B4P5T2 A0A1A9VCE1 A0A2M3YXS3 A0A1A9ZX37 A0A2M3ZZL4 W5J4R5 D3TR42 A0A182FH95 P35381 Q5XUA1 B3NNW0 D3W7E4 A0A171AYN4 A0A023F937 A0A0E3DQZ8 A0A0M4EW76 A0A1Q3FGZ3 A0A154PAS4 U5EZ82 B0W5F2 A0A1B6CGV5 T1PGB4 A0A0N0BIY2 J9JYX3 A0A411G6D1 A0A1B6FR19 A0A1L8EEH9 A0A1Y1LHJ4 R4FQG1 A0A1I8PLE2 A0A1L8EEN1 A0A087ZQI5 A0A1B6KWM9 A0A1L8EHL5 A0A146KQL2 A0A2A3E464 V9IFD6 B3MEJ0 A0A1A9WYG7 A0A3L8DC47 A0A0A9Z2F3 A0A0P5HII0 A0A164NWW6 A0A1D1ZI21 B4KPB8 K7IZG4 B4J414 A0A0L7QXV3 K7IZG5 B4LIV0
A0A212ELG1 A0A2W1BNE3 A0A2A4JQE0 D6WSI9 A0A2J7Q0B3 A0A034WCI4 A0A0A1WN96 A0A0K8TW13 A0A067QI29 S4VDE6 W8CCY0 A0A182N7Z7 Q7PHI8 A0A182XJA6 A0A182UN55 A0A182LBD1 A0A182HP56 A0A182MRL5 A0A182WAB9 A0A191UR49 A0A084W9J4 Q1HRQ7 A0A182JM63 A0A0V0G2J4 A0A182PHE7 A0A1L8E154 A0A1J1J483 A0A182Q0H6 A0A182RTQ3 A0A0L0CN23 A0A069DZ92 B4MQA2 A0A182K4A3 A0A336N1U6 A0A323TPU0 A0A423TIL1 J3JWJ5 A0A0J9RIM8 A0A1A9XAQ8 A0A1B0BN74 B4I8B0 A0A1W4VDL2 A0A0N7ZCJ7 T1DEX4 B4P5T2 A0A1A9VCE1 A0A2M3YXS3 A0A1A9ZX37 A0A2M3ZZL4 W5J4R5 D3TR42 A0A182FH95 P35381 Q5XUA1 B3NNW0 D3W7E4 A0A171AYN4 A0A023F937 A0A0E3DQZ8 A0A0M4EW76 A0A1Q3FGZ3 A0A154PAS4 U5EZ82 B0W5F2 A0A1B6CGV5 T1PGB4 A0A0N0BIY2 J9JYX3 A0A411G6D1 A0A1B6FR19 A0A1L8EEH9 A0A1Y1LHJ4 R4FQG1 A0A1I8PLE2 A0A1L8EEN1 A0A087ZQI5 A0A1B6KWM9 A0A1L8EHL5 A0A146KQL2 A0A2A3E464 V9IFD6 B3MEJ0 A0A1A9WYG7 A0A3L8DC47 A0A0A9Z2F3 A0A0P5HII0 A0A164NWW6 A0A1D1ZI21 B4KPB8 K7IZG4 B4J414 A0A0L7QXV3 K7IZG5 B4LIV0
EC Number
7.1.2.2
Pubmed
19121390
23622113
26354079
22118469
28756777
18362917
+ More
19820115 25348373 25830018 24845553 24495485 12364791 14747013 17210077 20966253 27538518 27142481 24438588 17204158 17510324 26108605 26334808 17994087 22516182 22936249 17550304 20920257 23761445 20353571 9540069 10731132 12537572 8500545 21384180 25474469 29136191 25315136 28004739 26823975 30249741 25401762 20075255
19820115 25348373 25830018 24845553 24495485 12364791 14747013 17210077 20966253 27538518 27142481 24438588 17204158 17510324 26108605 26334808 17994087 22516182 22936249 17550304 20920257 23761445 20353571 9540069 10731132 12537572 8500545 21384180 25474469 29136191 25315136 28004739 26823975 30249741 25401762 20075255
EMBL
BABH01018200
DQ311340
ABD36284.1
ODYU01005766
SOQ46956.1
GAIX01011955
+ More
JAA80605.1 RSAL01000096 RVE47723.1 KQ461073 KPJ09585.1 KQ459053 KPJ04023.1 AGBW02014073 OWR42307.1 KZ149982 PZC75791.1 NWSH01000823 PCG74029.1 KQ971354 EFA07428.1 NEVH01019983 PNF22023.1 GAKP01007449 JAC51503.1 GBXI01014419 JAC99872.1 GDHF01034079 GDHF01030438 GDHF01025175 JAI18235.1 JAI21876.1 JAI27139.1 KK853579 KDR06557.1 KC792298 AGO59887.1 GAMC01002911 JAC03645.1 AAAB01008900 EAA44524.2 APCN01002921 AXCM01001928 KU365938 KU932311 ANJ04654.1 APA33947.1 ATLV01021825 KE525324 KFB46888.1 DQ440037 CH477847 ABF18070.1 EAT35671.1 GECL01003752 JAP02372.1 GFDF01001782 JAV12302.1 CVRI01000070 CRL07216.1 AXCN02000909 JRES01000264 KNC32844.1 GBGD01001045 JAC87844.1 CH963849 EDW74291.1 UFQT01003371 SSX34933.1 MDVM02000029 PYZ99361.1 QCYY01001676 ROT76288.1 BT127613 AEE62575.1 CM002911 KMY95873.1 JXJN01017231 CH480824 EDW56835.1 GDRN01065788 JAI64640.1 GAMD01003346 JAA98244.1 CM000158 EDW91848.1 GGFM01000300 MBW21051.1 GGFK01000517 MBW33838.1 ADMH02002078 ETN59462.1 CCAG010006562 EZ423894 ADD20170.1 Y07894 AE013599 BT003592 AY737553 AAU84946.1 CH954179 EDV56693.1 GQ848643 ADC55251.1 GEMB01000492 JAS02635.1 GBBI01000721 JAC17991.1 KF818471 QKKF02010319 AIL26069.1 RZF44718.1 CP012524 ALC42287.1 GFDL01008243 JAV26802.1 KQ434846 KZC08318.1 GANO01001263 JAB58608.1 DS231842 EDS35272.1 GEDC01024675 JAS12623.1 KA647180 AFP61809.1 KQ435724 KOX78342.1 ABLF02029687 ABLF02029688 MH365641 QBB01450.1 GECZ01017155 JAS52614.1 GFDG01001745 JAV17054.1 GEZM01055159 GEZM01055158 JAV73104.1 ACPB03006347 GAHY01000414 JAA77096.1 GFDG01001663 JAV17136.1 GEBQ01024142 JAT15835.1 GFDG01000577 JAV18222.1 GDHC01019825 JAP98803.1 KZ288386 PBC26478.1 JR043517 AEY59785.1 CH902619 EDV37610.1 QOIP01000010 RLU17449.1 GBHO01007604 JAG36000.1 GDIQ01227542 JAK24183.1 LRGB01002793 KZS06308.1 GDJX01001320 JAT66616.1 CH933808 EDW09094.1 CH916367 EDW02620.1 KQ414699 KOC63448.1 CH940648 EDW61453.1
JAA80605.1 RSAL01000096 RVE47723.1 KQ461073 KPJ09585.1 KQ459053 KPJ04023.1 AGBW02014073 OWR42307.1 KZ149982 PZC75791.1 NWSH01000823 PCG74029.1 KQ971354 EFA07428.1 NEVH01019983 PNF22023.1 GAKP01007449 JAC51503.1 GBXI01014419 JAC99872.1 GDHF01034079 GDHF01030438 GDHF01025175 JAI18235.1 JAI21876.1 JAI27139.1 KK853579 KDR06557.1 KC792298 AGO59887.1 GAMC01002911 JAC03645.1 AAAB01008900 EAA44524.2 APCN01002921 AXCM01001928 KU365938 KU932311 ANJ04654.1 APA33947.1 ATLV01021825 KE525324 KFB46888.1 DQ440037 CH477847 ABF18070.1 EAT35671.1 GECL01003752 JAP02372.1 GFDF01001782 JAV12302.1 CVRI01000070 CRL07216.1 AXCN02000909 JRES01000264 KNC32844.1 GBGD01001045 JAC87844.1 CH963849 EDW74291.1 UFQT01003371 SSX34933.1 MDVM02000029 PYZ99361.1 QCYY01001676 ROT76288.1 BT127613 AEE62575.1 CM002911 KMY95873.1 JXJN01017231 CH480824 EDW56835.1 GDRN01065788 JAI64640.1 GAMD01003346 JAA98244.1 CM000158 EDW91848.1 GGFM01000300 MBW21051.1 GGFK01000517 MBW33838.1 ADMH02002078 ETN59462.1 CCAG010006562 EZ423894 ADD20170.1 Y07894 AE013599 BT003592 AY737553 AAU84946.1 CH954179 EDV56693.1 GQ848643 ADC55251.1 GEMB01000492 JAS02635.1 GBBI01000721 JAC17991.1 KF818471 QKKF02010319 AIL26069.1 RZF44718.1 CP012524 ALC42287.1 GFDL01008243 JAV26802.1 KQ434846 KZC08318.1 GANO01001263 JAB58608.1 DS231842 EDS35272.1 GEDC01024675 JAS12623.1 KA647180 AFP61809.1 KQ435724 KOX78342.1 ABLF02029687 ABLF02029688 MH365641 QBB01450.1 GECZ01017155 JAS52614.1 GFDG01001745 JAV17054.1 GEZM01055159 GEZM01055158 JAV73104.1 ACPB03006347 GAHY01000414 JAA77096.1 GFDG01001663 JAV17136.1 GEBQ01024142 JAT15835.1 GFDG01000577 JAV18222.1 GDHC01019825 JAP98803.1 KZ288386 PBC26478.1 JR043517 AEY59785.1 CH902619 EDV37610.1 QOIP01000010 RLU17449.1 GBHO01007604 JAG36000.1 GDIQ01227542 JAK24183.1 LRGB01002793 KZS06308.1 GDJX01001320 JAT66616.1 CH933808 EDW09094.1 CH916367 EDW02620.1 KQ414699 KOC63448.1 CH940648 EDW61453.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000053268
UP000007151
UP000218220
+ More
UP000007266 UP000235965 UP000027135 UP000075884 UP000007062 UP000076407 UP000075903 UP000075882 UP000075840 UP000075883 UP000075920 UP000030765 UP000008820 UP000075880 UP000075885 UP000183832 UP000075886 UP000075900 UP000037069 UP000007798 UP000075881 UP000247427 UP000283509 UP000092443 UP000092460 UP000001292 UP000192221 UP000002282 UP000078200 UP000092445 UP000000673 UP000092444 UP000069272 UP000000803 UP000008711 UP000291343 UP000092553 UP000076502 UP000002320 UP000095301 UP000053105 UP000007819 UP000015103 UP000095300 UP000005203 UP000242457 UP000007801 UP000091820 UP000279307 UP000076858 UP000009192 UP000002358 UP000001070 UP000053825 UP000008792
UP000007266 UP000235965 UP000027135 UP000075884 UP000007062 UP000076407 UP000075903 UP000075882 UP000075840 UP000075883 UP000075920 UP000030765 UP000008820 UP000075880 UP000075885 UP000183832 UP000075886 UP000075900 UP000037069 UP000007798 UP000075881 UP000247427 UP000283509 UP000092443 UP000092460 UP000001292 UP000192221 UP000002282 UP000078200 UP000092445 UP000000673 UP000092444 UP000069272 UP000000803 UP000008711 UP000291343 UP000092553 UP000076502 UP000002320 UP000095301 UP000053105 UP000007819 UP000015103 UP000095300 UP000005203 UP000242457 UP000007801 UP000091820 UP000279307 UP000076858 UP000009192 UP000002358 UP000001070 UP000053825 UP000008792
PRIDE
Interpro
IPR027417
P-loop_NTPase
+ More
IPR033732 ATP_synth_F1_a
IPR020003 ATPase_a/bsu_AS
IPR005294 ATP_synth_F1_asu
IPR000194 ATPase_F1/V1/A1_a/bsu_nucl-bd
IPR000793 ATP_synth_asu_C
IPR038376 ATP_synth_asu_C_sf
IPR036121 ATPase_F1/V1/A1_a/bsu_N_sf
IPR023366 ATP_synth_asu-like_sf
IPR004100 ATPase_F1/V1/A1_a/bsu_N
IPR033732 ATP_synth_F1_a
IPR020003 ATPase_a/bsu_AS
IPR005294 ATP_synth_F1_asu
IPR000194 ATPase_F1/V1/A1_a/bsu_nucl-bd
IPR000793 ATP_synth_asu_C
IPR038376 ATP_synth_asu_C_sf
IPR036121 ATPase_F1/V1/A1_a/bsu_N_sf
IPR023366 ATP_synth_asu-like_sf
IPR004100 ATPase_F1/V1/A1_a/bsu_N
Gene 3D
ProteinModelPortal
Q2F5T3
A0A2H1W1X7
S4P2L2
A0A3S2LJ09
A0A194QXA3
A0A194QEQ8
+ More
A0A212ELG1 A0A2W1BNE3 A0A2A4JQE0 D6WSI9 A0A2J7Q0B3 A0A034WCI4 A0A0A1WN96 A0A0K8TW13 A0A067QI29 S4VDE6 W8CCY0 A0A182N7Z7 Q7PHI8 A0A182XJA6 A0A182UN55 A0A182LBD1 A0A182HP56 A0A182MRL5 A0A182WAB9 A0A191UR49 A0A084W9J4 Q1HRQ7 A0A182JM63 A0A0V0G2J4 A0A182PHE7 A0A1L8E154 A0A1J1J483 A0A182Q0H6 A0A182RTQ3 A0A0L0CN23 A0A069DZ92 B4MQA2 A0A182K4A3 A0A336N1U6 A0A323TPU0 A0A423TIL1 J3JWJ5 A0A0J9RIM8 A0A1A9XAQ8 A0A1B0BN74 B4I8B0 A0A1W4VDL2 A0A0N7ZCJ7 T1DEX4 B4P5T2 A0A1A9VCE1 A0A2M3YXS3 A0A1A9ZX37 A0A2M3ZZL4 W5J4R5 D3TR42 A0A182FH95 P35381 Q5XUA1 B3NNW0 D3W7E4 A0A171AYN4 A0A023F937 A0A0E3DQZ8 A0A0M4EW76 A0A1Q3FGZ3 A0A154PAS4 U5EZ82 B0W5F2 A0A1B6CGV5 T1PGB4 A0A0N0BIY2 J9JYX3 A0A411G6D1 A0A1B6FR19 A0A1L8EEH9 A0A1Y1LHJ4 R4FQG1 A0A1I8PLE2 A0A1L8EEN1 A0A087ZQI5 A0A1B6KWM9 A0A1L8EHL5 A0A146KQL2 A0A2A3E464 V9IFD6 B3MEJ0 A0A1A9WYG7 A0A3L8DC47 A0A0A9Z2F3 A0A0P5HII0 A0A164NWW6 A0A1D1ZI21 B4KPB8 K7IZG4 B4J414 A0A0L7QXV3 K7IZG5 B4LIV0
A0A212ELG1 A0A2W1BNE3 A0A2A4JQE0 D6WSI9 A0A2J7Q0B3 A0A034WCI4 A0A0A1WN96 A0A0K8TW13 A0A067QI29 S4VDE6 W8CCY0 A0A182N7Z7 Q7PHI8 A0A182XJA6 A0A182UN55 A0A182LBD1 A0A182HP56 A0A182MRL5 A0A182WAB9 A0A191UR49 A0A084W9J4 Q1HRQ7 A0A182JM63 A0A0V0G2J4 A0A182PHE7 A0A1L8E154 A0A1J1J483 A0A182Q0H6 A0A182RTQ3 A0A0L0CN23 A0A069DZ92 B4MQA2 A0A182K4A3 A0A336N1U6 A0A323TPU0 A0A423TIL1 J3JWJ5 A0A0J9RIM8 A0A1A9XAQ8 A0A1B0BN74 B4I8B0 A0A1W4VDL2 A0A0N7ZCJ7 T1DEX4 B4P5T2 A0A1A9VCE1 A0A2M3YXS3 A0A1A9ZX37 A0A2M3ZZL4 W5J4R5 D3TR42 A0A182FH95 P35381 Q5XUA1 B3NNW0 D3W7E4 A0A171AYN4 A0A023F937 A0A0E3DQZ8 A0A0M4EW76 A0A1Q3FGZ3 A0A154PAS4 U5EZ82 B0W5F2 A0A1B6CGV5 T1PGB4 A0A0N0BIY2 J9JYX3 A0A411G6D1 A0A1B6FR19 A0A1L8EEH9 A0A1Y1LHJ4 R4FQG1 A0A1I8PLE2 A0A1L8EEN1 A0A087ZQI5 A0A1B6KWM9 A0A1L8EHL5 A0A146KQL2 A0A2A3E464 V9IFD6 B3MEJ0 A0A1A9WYG7 A0A3L8DC47 A0A0A9Z2F3 A0A0P5HII0 A0A164NWW6 A0A1D1ZI21 B4KPB8 K7IZG4 B4J414 A0A0L7QXV3 K7IZG5 B4LIV0
PDB
2W6J
E-value=0,
Score=2215
Ontologies
PATHWAY
GO
Topology
Subcellular location
Cell membrane
Mitochondrion inner membrane
Mitochondrion inner membrane
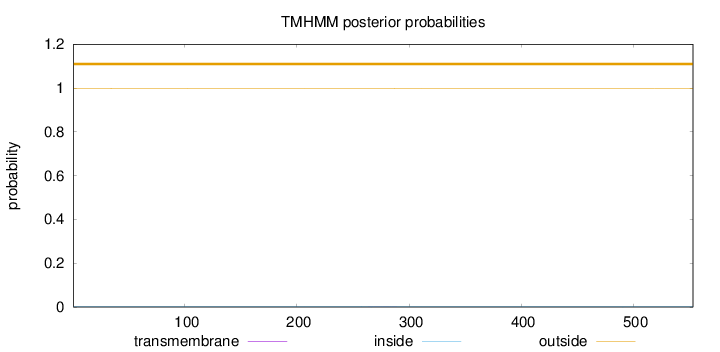
Length:
553
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02359
Exp number, first 60 AAs:
0.00165
Total prob of N-in:
0.00157
outside
1 - 553
Population Genetic Test Statistics
Pi
264.121357
Theta
209.321834
Tajima's D
1.042591
CLR
0.001077
CSRT
0.666816659167042
Interpretation
Uncertain
