Gene
KWMTBOMO11350
Pre Gene Modal
BGIBMGA001967
Annotation
PREDICTED:_PX_domain-containing_protein_kinase-like_protein_[Bombyx_mori]
Full name
PX domain-containing protein kinase-like protein
Alternative Name
Modulator of Na,K-ATPase
Location in the cell
Nuclear Reliability : 2.223
Sequence
CDS
ATGGCTTTGTGGCAGAATGCGTCGTCCGTCATATGGAGATTAGACGACACAGCGAAAATTACATGTTTCATAGAAAATGCTCAGAACATTAACAAACACACCGAATACATAATTAGGGTTCAAAGAGGCCCCGTCAAAGAAAATATGTGGAATGTATCTCGTAGATATAGTGATTTTGCATCGCTACACTCATTACTGCTGAAATCCAATATAGATTTGCCTCTCCCACCTAAAAAACTCATAGGAAATATGCAGCCTTCATTTATCGCAGAAAGACAAGTGGCATTGCAGAACTATTTAAATGAAGTGCTGAAACATCAGATATTAGCATTGTCATTGTCAGTGAGAAGTTTTCTAGATCCAAACAACTATTCATTAGCCATCTCAGAGCAATCTCTGCAGACCGTGTCAATAGCCTTGAGGGGAGACTTGCGCTATGAACTAAAAGGACCCATTGCTGATATTGGATGGAGAATAAGGAAACATTATTTTTTGATGTCCGATATTAGTAGCAAGACTAATTGTTTACTTTCGTATCAGCACTACGGTCCGGATAGATATCTCTGCGATAAAGACTTGCAGACTGCCTTCAAGAGCCTACAAAATCTTTCGCACCCTTATATAGACCAAATATTAGCTATACATAATTTAGAAAATGGTGCCTACGTTATTAGAAGGATACACGAGAACGGAAGTCTGAGAGATATATTATACGGTACCGAATACAGTAAACATTATTTAATGAAGTATGGCAACCCGAAAATAAGGAAACCATTCACGACCGGACAAATCGCACACTATGGTTATCAGATATTGCAAGCAATAAAGTTTCTACACGAAAAAGGTCTACCACACGGTCACATCCATCCCGGCAACATCGCAATAGAGAACCAAAGAGCTCTGCTAATGGAGGTCGAGAATTTTGTTATCGGCGTGCCCTGTTTGTACCGGCCCTATCTACTCGAACTGCGGAGGGTGACGACCCCCGAAGACATCGACGTATACTGCTTCGGCAGAACCTTGTACGAGATGACGTTCGCCATCGCACCGGAGGACCCCCACTGCGATTACTTCCCCGACGGAATCAATATGGATTTAGAATCAGTATTACACATGTGCCTTTCAAGTCACGCTTGCAAGCACGGCATGCCGACAGTAGACAGTCTACTCCGGCACCCGTTCTTCTGCCAAGCGCCCCTCAACGGCCTCACGGTGGCGCCGCGCGACGAGAAGATCCACCTCAAGTTCCCCATACAGCTGAAGGAAGAGTTGAAGGCCGCCGTCTCTAGCGCCGAAGCGAGATTAAAACGAGAACAGAAACTGGTGCGAAGCGCAAAGAGAGAAGTCAGGATCCAAGAAATACTCTCTTCAGAAGAGGAAATGAAGAAACAGAAAAAAAGAATGAAAAAGCGCGAGAGCATCTGGAAGTCGACGTCGTCGCTGGCGGACACGGTGCGGAGCGCCAGCACGGCCTCGTCCCCCTCGCCGCCCGCGCCCCGCGAGCGCACCCCGCCGCCCCCGCCACAAGGCGCAGACATGGGTGCCGCGGAGGGAGCGCCGCCCCCCGCCGGCCGCTCCGCGCTGCTGCACGCCATCTGCGCCTTCGACAAGTCCAGACTCGCTCGCGTATCGACTCGGTGA
Protein
MALWQNASSVIWRLDDTAKITCFIENAQNINKHTEYIIRVQRGPVKENMWNVSRRYSDFASLHSLLLKSNIDLPLPPKKLIGNMQPSFIAERQVALQNYLNEVLKHQILALSLSVRSFLDPNNYSLAISEQSLQTVSIALRGDLRYELKGPIADIGWRIRKHYFLMSDISSKTNCLLSYQHYGPDRYLCDKDLQTAFKSLQNLSHPYIDQILAIHNLENGAYVIRRIHENGSLRDILYGTEYSKHYLMKYGNPKIRKPFTTGQIAHYGYQILQAIKFLHEKGLPHGHIHPGNIAIENQRALLMEVENFVIGVPCLYRPYLLELRRVTTPEDIDVYCFGRTLYEMTFAIAPEDPHCDYFPDGINMDLESVLHMCLSSHACKHGMPTVDSLLRHPFFCQAPLNGLTVAPRDEKIHLKFPIQLKEELKAAVSSAEARLKREQKLVRSAKREVRIQEILSSEEEMKKQKKRMKKRESIWKSTSSLADTVRSASTASSPSPPAPRERTPPPPPQGADMGAAEGAPPPAGRSALLHAICAFDKSRLARVSTR
Summary
Description
Binds to and modulates brain Na,K-ATPase subunits ATP1B1 and ATP1B3 and may thereby participate in the regulation of electrical excitability and synaptic transmission. May not display kinase activity.
Similarity
Belongs to the protein kinase superfamily.
Keywords
Actin-binding
Alternative splicing
Cell membrane
Complete proteome
Cytoplasm
Membrane
Reference proteome
Feature
chain PX domain-containing protein kinase-like protein
splice variant In isoform 3.
splice variant In isoform 3.
Uniprot
H9IXI4
A0A2H1W1P7
A0A2A4JQQ0
A0A2W1BSQ6
A0A194QWA4
A0A3S2NXD8
+ More
A0A212ELF5 A0A194QF29 A0A2J7PZP6 A0A2J7PZN0 D6WQ78 A0A067QV47 A0A1Y1LI12 V9IAU7 A0A2A3ER94 A0A088A0Q9 A0A0C9QC73 A0A0C9RGN4 A0A0J7KQX6 A0A0C9Q9W3 A0A232F530 K7J816 A0A1W4WMQ9 E9IVE1 A0A151XHQ6 F4X7A4 A0A195FGQ5 E1ZW46 E2C009 A0A026WUI1 A0A195B1E4 A0A158ND63 A0A0L7QX31 A0A151ICB2 A0A195DJF0 U4UJH9 A0A1B6CZC3 N6URM8 A0A1B6G353 A0A3B3QXH9 A0A3Q1B738 A0A3P9A4A9 A0A3Q1CCD9 H2MYY7 A0A2D0PLG0 A0A3Q1B4P4 A0A0B8RU11 A0A3P8UCA9 A0A3B4VAU0 W5UJE9 E6ZGF0 A0A2T7NTX1 G1KLK4 A0A1U8C6N2 A0A3Q0CXX7 A0A1U7QYL6 A0A3Q0D3E4 A0A2U9BE91 Q6DFG1 A0A147AH73 A0A3B4DG86 A0A087XLM7 A0A3Q2VL46 U3F7W1 A0A1W4Z8Q0 A0A3Q1IIN0 A0A2D4JWI2 A0A0L8G5U9 A0A1U7QN72 A0A3B3XQ31 A0A3B3YYT2 A0A3P8WQH2 A0A096MDR5 A0A3Q0D1E0 A0A0L8G5U6 I3IV85 A0A1W4ZHV7 A0A0N8K0L4 A0A1B6KY41 A0A3B4H5P6 A0A0F8CH29 A0A1V4JIC7 A0A2D4P8G2 Q8BX57-2 A0A2I4BSP6 W5KBV6 F1SGJ0 A0A3P9NX70 A0A1V4JJJ7 A0A1V4JIF3 A0A1S3K5R6 A0A2Y9N5S6 A0A2Y9N2L0 A0A2Y9NGP3 A0A2U4CCC9 A0A2U4CCC3 A0A2U4CCF3 A0A226PH43 A0A2Y9NAG6 A0A2K6RMV6
A0A212ELF5 A0A194QF29 A0A2J7PZP6 A0A2J7PZN0 D6WQ78 A0A067QV47 A0A1Y1LI12 V9IAU7 A0A2A3ER94 A0A088A0Q9 A0A0C9QC73 A0A0C9RGN4 A0A0J7KQX6 A0A0C9Q9W3 A0A232F530 K7J816 A0A1W4WMQ9 E9IVE1 A0A151XHQ6 F4X7A4 A0A195FGQ5 E1ZW46 E2C009 A0A026WUI1 A0A195B1E4 A0A158ND63 A0A0L7QX31 A0A151ICB2 A0A195DJF0 U4UJH9 A0A1B6CZC3 N6URM8 A0A1B6G353 A0A3B3QXH9 A0A3Q1B738 A0A3P9A4A9 A0A3Q1CCD9 H2MYY7 A0A2D0PLG0 A0A3Q1B4P4 A0A0B8RU11 A0A3P8UCA9 A0A3B4VAU0 W5UJE9 E6ZGF0 A0A2T7NTX1 G1KLK4 A0A1U8C6N2 A0A3Q0CXX7 A0A1U7QYL6 A0A3Q0D3E4 A0A2U9BE91 Q6DFG1 A0A147AH73 A0A3B4DG86 A0A087XLM7 A0A3Q2VL46 U3F7W1 A0A1W4Z8Q0 A0A3Q1IIN0 A0A2D4JWI2 A0A0L8G5U9 A0A1U7QN72 A0A3B3XQ31 A0A3B3YYT2 A0A3P8WQH2 A0A096MDR5 A0A3Q0D1E0 A0A0L8G5U6 I3IV85 A0A1W4ZHV7 A0A0N8K0L4 A0A1B6KY41 A0A3B4H5P6 A0A0F8CH29 A0A1V4JIC7 A0A2D4P8G2 Q8BX57-2 A0A2I4BSP6 W5KBV6 F1SGJ0 A0A3P9NX70 A0A1V4JJJ7 A0A1V4JIF3 A0A1S3K5R6 A0A2Y9N5S6 A0A2Y9N2L0 A0A2Y9NGP3 A0A2U4CCC9 A0A2U4CCC3 A0A2U4CCF3 A0A226PH43 A0A2Y9NAG6 A0A2K6RMV6
Pubmed
19121390
28756777
26354079
22118469
18362917
19820115
+ More
24845553 28004739 28648823 20075255 21282665 21719571 20798317 24508170 30249741 21347285 23537049 29240929 25069045 17554307 25476704 23127152 28071753 23915248 24487278 25186727 25835551 16135750 16141072 15489334 21183079 25329095 30723633 24621616 25362486
24845553 28004739 28648823 20075255 21282665 21719571 20798317 24508170 30249741 21347285 23537049 29240929 25069045 17554307 25476704 23127152 28071753 23915248 24487278 25186727 25835551 16135750 16141072 15489334 21183079 25329095 30723633 24621616 25362486
EMBL
BABH01018200
BABH01018201
ODYU01005766
SOQ46957.1
NWSH01000823
PCG74028.1
+ More
KZ149982 PZC75790.1 KQ461073 KPJ09584.1 RSAL01000034 RVE51423.1 AGBW02014073 OWR42308.1 KQ459053 KPJ04024.1 NEVH01020333 PNF21792.1 PNF21791.1 KQ971354 EFA06114.1 KK852981 KDR12927.1 GEZM01054974 JAV73289.1 JR037082 AEY57571.1 KZ288201 PBC33561.1 GBYB01000884 JAG70651.1 GBYB01012267 JAG82034.1 LBMM01004213 KMQ92696.1 GBYB01000004 JAG69771.1 NNAY01000991 OXU25588.1 GL766194 EFZ15493.1 KQ982116 KYQ59944.1 GL888828 EGI57716.1 KQ981560 KYN39865.1 GL434776 EFN74555.1 GL451712 EFN78705.1 KK107102 QOIP01000005 EZA59613.1 RLU22381.1 KQ976690 KYM78107.1 ADTU01012423 KQ414705 KOC63177.1 KQ978060 KYM97730.1 KQ980800 KYN12942.1 KB632331 ERL92628.1 GEDC01018573 JAS18725.1 APGK01019272 KB740098 ENN81392.1 GECZ01012948 GECZ01003675 JAS56821.1 JAS66094.1 GBSH01002145 JAG66881.1 JT417147 AHH42274.1 FQ310506 CBN80667.1 PZQS01000009 PVD24611.1 CP026248 AWP02293.1 BC076777 AAH76777.1 GCES01008524 JAR77799.1 AYCK01023250 GAEP01000799 JAB54022.1 IACL01013140 LAB00829.1 KQ423675 KOF72412.1 KOF72411.1 AERX01018480 AERX01018481 AERX01018482 JARO02002523 KPP72465.1 GEBQ01023618 JAT16359.1 KQ040939 KKF32833.1 LSYS01007300 OPJ71949.1 IACN01057743 LAB53569.1 DQ095196 DQ095197 AK048910 AK152058 AK152789 AK153067 AK156146 AK157364 AK169565 AK170361 AK150413 AK150337 BC016131 AAH16131.1 AAZ04665.1 BAC33489.1 BAE29537.1 BAE34065.1 BAE41231.1 BAE41744.1 AEMK02000086 DQIR01314807 HDC70285.1 OPJ71947.1 OPJ71948.1 AWGT02000081 OXB78707.1
KZ149982 PZC75790.1 KQ461073 KPJ09584.1 RSAL01000034 RVE51423.1 AGBW02014073 OWR42308.1 KQ459053 KPJ04024.1 NEVH01020333 PNF21792.1 PNF21791.1 KQ971354 EFA06114.1 KK852981 KDR12927.1 GEZM01054974 JAV73289.1 JR037082 AEY57571.1 KZ288201 PBC33561.1 GBYB01000884 JAG70651.1 GBYB01012267 JAG82034.1 LBMM01004213 KMQ92696.1 GBYB01000004 JAG69771.1 NNAY01000991 OXU25588.1 GL766194 EFZ15493.1 KQ982116 KYQ59944.1 GL888828 EGI57716.1 KQ981560 KYN39865.1 GL434776 EFN74555.1 GL451712 EFN78705.1 KK107102 QOIP01000005 EZA59613.1 RLU22381.1 KQ976690 KYM78107.1 ADTU01012423 KQ414705 KOC63177.1 KQ978060 KYM97730.1 KQ980800 KYN12942.1 KB632331 ERL92628.1 GEDC01018573 JAS18725.1 APGK01019272 KB740098 ENN81392.1 GECZ01012948 GECZ01003675 JAS56821.1 JAS66094.1 GBSH01002145 JAG66881.1 JT417147 AHH42274.1 FQ310506 CBN80667.1 PZQS01000009 PVD24611.1 CP026248 AWP02293.1 BC076777 AAH76777.1 GCES01008524 JAR77799.1 AYCK01023250 GAEP01000799 JAB54022.1 IACL01013140 LAB00829.1 KQ423675 KOF72412.1 KOF72411.1 AERX01018480 AERX01018481 AERX01018482 JARO02002523 KPP72465.1 GEBQ01023618 JAT16359.1 KQ040939 KKF32833.1 LSYS01007300 OPJ71949.1 IACN01057743 LAB53569.1 DQ095196 DQ095197 AK048910 AK152058 AK152789 AK153067 AK156146 AK157364 AK169565 AK170361 AK150413 AK150337 BC016131 AAH16131.1 AAZ04665.1 BAC33489.1 BAE29537.1 BAE34065.1 BAE41231.1 BAE41744.1 AEMK02000086 DQIR01314807 HDC70285.1 OPJ71947.1 OPJ71948.1 AWGT02000081 OXB78707.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000283053
UP000007151
UP000053268
+ More
UP000235965 UP000007266 UP000027135 UP000242457 UP000005203 UP000036403 UP000215335 UP000002358 UP000192223 UP000075809 UP000007755 UP000078541 UP000000311 UP000008237 UP000053097 UP000279307 UP000078540 UP000005205 UP000053825 UP000078542 UP000078492 UP000030742 UP000019118 UP000261540 UP000257160 UP000265140 UP000001038 UP000221080 UP000265080 UP000261420 UP000245119 UP000001646 UP000189706 UP000246464 UP000261440 UP000028760 UP000264840 UP000192224 UP000265040 UP000053454 UP000261480 UP000265120 UP000005207 UP000034805 UP000261460 UP000190648 UP000000589 UP000192220 UP000018467 UP000008227 UP000242638 UP000085678 UP000248483 UP000245320 UP000198419 UP000233200
UP000235965 UP000007266 UP000027135 UP000242457 UP000005203 UP000036403 UP000215335 UP000002358 UP000192223 UP000075809 UP000007755 UP000078541 UP000000311 UP000008237 UP000053097 UP000279307 UP000078540 UP000005205 UP000053825 UP000078542 UP000078492 UP000030742 UP000019118 UP000261540 UP000257160 UP000265140 UP000001038 UP000221080 UP000265080 UP000261420 UP000245119 UP000001646 UP000189706 UP000246464 UP000261440 UP000028760 UP000264840 UP000192224 UP000265040 UP000053454 UP000261480 UP000265120 UP000005207 UP000034805 UP000261460 UP000190648 UP000000589 UP000192220 UP000018467 UP000008227 UP000242638 UP000085678 UP000248483 UP000245320 UP000198419 UP000233200
Interpro
Gene 3D
ProteinModelPortal
H9IXI4
A0A2H1W1P7
A0A2A4JQQ0
A0A2W1BSQ6
A0A194QWA4
A0A3S2NXD8
+ More
A0A212ELF5 A0A194QF29 A0A2J7PZP6 A0A2J7PZN0 D6WQ78 A0A067QV47 A0A1Y1LI12 V9IAU7 A0A2A3ER94 A0A088A0Q9 A0A0C9QC73 A0A0C9RGN4 A0A0J7KQX6 A0A0C9Q9W3 A0A232F530 K7J816 A0A1W4WMQ9 E9IVE1 A0A151XHQ6 F4X7A4 A0A195FGQ5 E1ZW46 E2C009 A0A026WUI1 A0A195B1E4 A0A158ND63 A0A0L7QX31 A0A151ICB2 A0A195DJF0 U4UJH9 A0A1B6CZC3 N6URM8 A0A1B6G353 A0A3B3QXH9 A0A3Q1B738 A0A3P9A4A9 A0A3Q1CCD9 H2MYY7 A0A2D0PLG0 A0A3Q1B4P4 A0A0B8RU11 A0A3P8UCA9 A0A3B4VAU0 W5UJE9 E6ZGF0 A0A2T7NTX1 G1KLK4 A0A1U8C6N2 A0A3Q0CXX7 A0A1U7QYL6 A0A3Q0D3E4 A0A2U9BE91 Q6DFG1 A0A147AH73 A0A3B4DG86 A0A087XLM7 A0A3Q2VL46 U3F7W1 A0A1W4Z8Q0 A0A3Q1IIN0 A0A2D4JWI2 A0A0L8G5U9 A0A1U7QN72 A0A3B3XQ31 A0A3B3YYT2 A0A3P8WQH2 A0A096MDR5 A0A3Q0D1E0 A0A0L8G5U6 I3IV85 A0A1W4ZHV7 A0A0N8K0L4 A0A1B6KY41 A0A3B4H5P6 A0A0F8CH29 A0A1V4JIC7 A0A2D4P8G2 Q8BX57-2 A0A2I4BSP6 W5KBV6 F1SGJ0 A0A3P9NX70 A0A1V4JJJ7 A0A1V4JIF3 A0A1S3K5R6 A0A2Y9N5S6 A0A2Y9N2L0 A0A2Y9NGP3 A0A2U4CCC9 A0A2U4CCC3 A0A2U4CCF3 A0A226PH43 A0A2Y9NAG6 A0A2K6RMV6
A0A212ELF5 A0A194QF29 A0A2J7PZP6 A0A2J7PZN0 D6WQ78 A0A067QV47 A0A1Y1LI12 V9IAU7 A0A2A3ER94 A0A088A0Q9 A0A0C9QC73 A0A0C9RGN4 A0A0J7KQX6 A0A0C9Q9W3 A0A232F530 K7J816 A0A1W4WMQ9 E9IVE1 A0A151XHQ6 F4X7A4 A0A195FGQ5 E1ZW46 E2C009 A0A026WUI1 A0A195B1E4 A0A158ND63 A0A0L7QX31 A0A151ICB2 A0A195DJF0 U4UJH9 A0A1B6CZC3 N6URM8 A0A1B6G353 A0A3B3QXH9 A0A3Q1B738 A0A3P9A4A9 A0A3Q1CCD9 H2MYY7 A0A2D0PLG0 A0A3Q1B4P4 A0A0B8RU11 A0A3P8UCA9 A0A3B4VAU0 W5UJE9 E6ZGF0 A0A2T7NTX1 G1KLK4 A0A1U8C6N2 A0A3Q0CXX7 A0A1U7QYL6 A0A3Q0D3E4 A0A2U9BE91 Q6DFG1 A0A147AH73 A0A3B4DG86 A0A087XLM7 A0A3Q2VL46 U3F7W1 A0A1W4Z8Q0 A0A3Q1IIN0 A0A2D4JWI2 A0A0L8G5U9 A0A1U7QN72 A0A3B3XQ31 A0A3B3YYT2 A0A3P8WQH2 A0A096MDR5 A0A3Q0D1E0 A0A0L8G5U6 I3IV85 A0A1W4ZHV7 A0A0N8K0L4 A0A1B6KY41 A0A3B4H5P6 A0A0F8CH29 A0A1V4JIC7 A0A2D4P8G2 Q8BX57-2 A0A2I4BSP6 W5KBV6 F1SGJ0 A0A3P9NX70 A0A1V4JJJ7 A0A1V4JIF3 A0A1S3K5R6 A0A2Y9N5S6 A0A2Y9N2L0 A0A2Y9NGP3 A0A2U4CCC9 A0A2U4CCC3 A0A2U4CCF3 A0A226PH43 A0A2Y9NAG6 A0A2K6RMV6
PDB
5GW1
E-value=1.54999e-07,
Score=134
Ontologies
GO
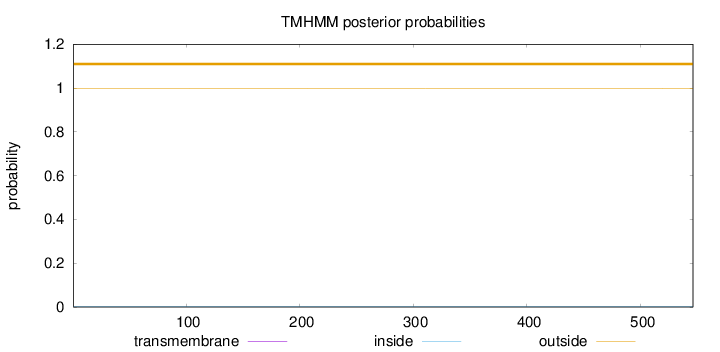
Topology
Subcellular location
Cytoplasm
Cell membrane
Cell membrane
Length:
546
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00171
Exp number, first 60 AAs:
5e-05
Total prob of N-in:
0.00180
outside
1 - 546
Population Genetic Test Statistics
Pi
268.932138
Theta
170.982455
Tajima's D
1.29044
CLR
0.240662
CSRT
0.738013099345033
Interpretation
Uncertain
