Gene
KWMTBOMO11345
Pre Gene Modal
BGIBMGA001969
Annotation
PREDICTED:_A_disintegrin_and_metalloproteinase_with_thrombospondin_motifs_16_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.119
Sequence
CDS
ATGGTCTGCAGCAAATACAAAGAACGCGTCAGAAGATTATCAGGCCTCGGCATGCAGATATCGCCGGCTCTAGAGGACCCGGACCGGCCGTGCCGCATCGCGTGCCAGGACGAGCGCGTCGCGCACCGCTTCTACTTGGTCAACGGGCACGAGGGCTGGTTCCCGCTGGGCACGGCCTGCGGGCACAACAACGCCTCCTACTGCGTCAGCGGGAAGTGCTTGGAGTTCGGCGGAGACAACACCCCGGTCCCGGAGACGGTGTACGCGCTGCCGCTGCTGCCGCGCCGCGTCACCCGCAGCCTCACCGCGCGGATCGACGTCTCGCACCTCGACGACCTCATCACCGCGCTCAACCTCACACATACCCCGGAGGCTCACATAACGTACTACGACAGTATACCAGACAACGTCGAAGTGGACTTCAACAATCCCATACACATATCTCCGGAGGACACGTTATTGAGGAAAAAACAGAGACCGACGTGGGACTGA
Protein
MVCSKYKERVRRLSGLGMQISPALEDPDRPCRIACQDERVAHRFYLVNGHEGWFPLGTACGHNNASYCVSGKCLEFGGDNTPVPETVYALPLLPRRVTRSLTARIDVSHLDDLITALNLTHTPEAHITYYDSIPDNVEVDFNNPIHISPEDTLLRKKQRPTWD
Summary
Uniprot
H9IXI6
A0A437ARJ2
A0A194QEY4
A0A2W1BSJ3
A0A194QW27
A0A2A4IYQ3
+ More
A0A212F8C6 U4UKR6 N6SSR5 A0A067RCW6 D6WRW7 A0A2J7R4U0 A0A1Y1LAA0 A0A1Y1LDZ5 A0A1B6EY69 A0A2J7R4T8 E0VAR6 A0A1S4FP70 A0A182H310 A0A182JFI6 Q16TY3 A0A2Y9D2Y1 A0A182XEH7 A0A182N1Y1 A0A1B0D865 A0A182WM27 A0A1A9ZX31 A0A182HQM0 A0A182UJG2 Q7Q7Y1 A0A1B0GKN0 A0A0A9XI82 A0A0A9XN21 A0A0L0CKU5 A0A0A9XCR4 B4I8B2 Q8T3M5 A0A2S2QNX1 A0A034V3Q9 A0A2S2P9A4 W8APV3 A0A1B0EUB2 A0A2S2RB68 A0A1A9VA32 A0A1A9XAQ9 A0A1B0BN75 J9JTN6 A0A1A9WYG6 B4P5T4 B4J412 A0A2S2PIE9 A0A336KUH1 B4MQA0 A0A3B0IY20 A0A0K8VW49 A0A336MQA6 A0A2H8TN60 A0A1I8PMI4 A0A0A1X1I4 B4LIV2 A0A0Q9W4L5 A0A1W4VQL9 Q8T458 A0A0M3QVE7 B4GAH9 A0A0K8U7W2 Q291N2 B3NNW2 A0A0J9RJ86 B4QHN1 Q9W1Z6 B4KPC0 A0A3B0JHX5 A0A3B0IYE1 B3MEJ2 A0A182YR46 A0A182S8R9
A0A212F8C6 U4UKR6 N6SSR5 A0A067RCW6 D6WRW7 A0A2J7R4U0 A0A1Y1LAA0 A0A1Y1LDZ5 A0A1B6EY69 A0A2J7R4T8 E0VAR6 A0A1S4FP70 A0A182H310 A0A182JFI6 Q16TY3 A0A2Y9D2Y1 A0A182XEH7 A0A182N1Y1 A0A1B0D865 A0A182WM27 A0A1A9ZX31 A0A182HQM0 A0A182UJG2 Q7Q7Y1 A0A1B0GKN0 A0A0A9XI82 A0A0A9XN21 A0A0L0CKU5 A0A0A9XCR4 B4I8B2 Q8T3M5 A0A2S2QNX1 A0A034V3Q9 A0A2S2P9A4 W8APV3 A0A1B0EUB2 A0A2S2RB68 A0A1A9VA32 A0A1A9XAQ9 A0A1B0BN75 J9JTN6 A0A1A9WYG6 B4P5T4 B4J412 A0A2S2PIE9 A0A336KUH1 B4MQA0 A0A3B0IY20 A0A0K8VW49 A0A336MQA6 A0A2H8TN60 A0A1I8PMI4 A0A0A1X1I4 B4LIV2 A0A0Q9W4L5 A0A1W4VQL9 Q8T458 A0A0M3QVE7 B4GAH9 A0A0K8U7W2 Q291N2 B3NNW2 A0A0J9RJ86 B4QHN1 Q9W1Z6 B4KPC0 A0A3B0JHX5 A0A3B0IYE1 B3MEJ2 A0A182YR46 A0A182S8R9
Pubmed
19121390
26354079
28756777
22118469
23537049
24845553
+ More
18362917 19820115 28004739 20566863 26483478 17510324 12364791 14747013 17210077 25401762 26108605 17994087 25348373 24495485 17550304 25830018 18057021 15632085 23185243 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25244985
18362917 19820115 28004739 20566863 26483478 17510324 12364791 14747013 17210077 25401762 26108605 17994087 25348373 24495485 17550304 25830018 18057021 15632085 23185243 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25244985
EMBL
BABH01018208
BABH01018209
BABH01018210
RSAL01004745
RVE40099.1
KQ459053
+ More
KPJ04103.1 KZ149913 PZC78038.1 KQ461073 KPJ09514.1 NWSH01005320 PCG64292.1 AGBW02009752 OWR49996.1 KB632370 ERL93707.1 APGK01058444 APGK01058445 APGK01058446 KB741288 ENN70714.1 KK852718 KDR17787.1 KQ971351 EFA06592.2 NEVH01007402 PNF35841.1 GEZM01061283 JAV70569.1 GEZM01061282 JAV70570.1 GECZ01026783 JAS42986.1 PNF35840.1 DS235014 EEB10472.1 JXUM01024343 JXUM01024344 JXUM01024345 KQ560687 KXJ81277.1 CH477637 EAT37983.1 AJVK01027234 APCN01004658 AAAB01008948 EAA10377.4 AJWK01030743 AJWK01030744 GBHO01025096 GBHO01025091 JAG18508.1 JAG18513.1 GBHO01025094 JAG18510.1 JRES01000264 KNC32866.1 GBHO01025092 JAG18512.1 CH480824 EDW56837.1 AY094759 AAM11112.1 GGMS01010263 MBY79466.1 GAKP01022185 GAKP01022184 GAKP01022181 JAC36767.1 GGMR01013424 MBY26043.1 GAMC01018498 JAB88057.1 CCAG010013080 GGMS01018001 MBY87204.1 JXJN01017228 ABLF02023476 CM000158 EDW91850.2 CH916367 EDW02618.1 GGMR01016602 MBY29221.1 UFQS01000762 UFQT01000762 SSX06703.1 SSX27049.1 CH963849 EDW74289.1 OUUW01000001 SPP72824.1 GDHF01009210 JAI43104.1 UFQT01000946 SSX28158.1 GFXV01003798 MBW15603.1 GBXI01009335 JAD04957.1 CH940648 EDW61455.1 KRF79993.1 KRF79994.1 KRF79995.1 KRF79996.1 AY089340 AAL90078.1 CP012524 ALC42289.1 CH479181 EDW31931.1 GDHF01029869 JAI22445.1 CM000071 EAL25080.2 KRT01835.1 KRT01836.1 CH954179 EDV56695.2 CM002911 KMY95877.1 CM000362 EDX08260.1 AE013599 AAF46905.2 AFH08222.1 CH933808 EDW09096.2 SPP72826.1 SPP72825.1 CH902619 EDV37612.1
KPJ04103.1 KZ149913 PZC78038.1 KQ461073 KPJ09514.1 NWSH01005320 PCG64292.1 AGBW02009752 OWR49996.1 KB632370 ERL93707.1 APGK01058444 APGK01058445 APGK01058446 KB741288 ENN70714.1 KK852718 KDR17787.1 KQ971351 EFA06592.2 NEVH01007402 PNF35841.1 GEZM01061283 JAV70569.1 GEZM01061282 JAV70570.1 GECZ01026783 JAS42986.1 PNF35840.1 DS235014 EEB10472.1 JXUM01024343 JXUM01024344 JXUM01024345 KQ560687 KXJ81277.1 CH477637 EAT37983.1 AJVK01027234 APCN01004658 AAAB01008948 EAA10377.4 AJWK01030743 AJWK01030744 GBHO01025096 GBHO01025091 JAG18508.1 JAG18513.1 GBHO01025094 JAG18510.1 JRES01000264 KNC32866.1 GBHO01025092 JAG18512.1 CH480824 EDW56837.1 AY094759 AAM11112.1 GGMS01010263 MBY79466.1 GAKP01022185 GAKP01022184 GAKP01022181 JAC36767.1 GGMR01013424 MBY26043.1 GAMC01018498 JAB88057.1 CCAG010013080 GGMS01018001 MBY87204.1 JXJN01017228 ABLF02023476 CM000158 EDW91850.2 CH916367 EDW02618.1 GGMR01016602 MBY29221.1 UFQS01000762 UFQT01000762 SSX06703.1 SSX27049.1 CH963849 EDW74289.1 OUUW01000001 SPP72824.1 GDHF01009210 JAI43104.1 UFQT01000946 SSX28158.1 GFXV01003798 MBW15603.1 GBXI01009335 JAD04957.1 CH940648 EDW61455.1 KRF79993.1 KRF79994.1 KRF79995.1 KRF79996.1 AY089340 AAL90078.1 CP012524 ALC42289.1 CH479181 EDW31931.1 GDHF01029869 JAI22445.1 CM000071 EAL25080.2 KRT01835.1 KRT01836.1 CH954179 EDV56695.2 CM002911 KMY95877.1 CM000362 EDX08260.1 AE013599 AAF46905.2 AFH08222.1 CH933808 EDW09096.2 SPP72826.1 SPP72825.1 CH902619 EDV37612.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000053240
UP000218220
UP000007151
+ More
UP000030742 UP000019118 UP000027135 UP000007266 UP000235965 UP000009046 UP000069940 UP000249989 UP000075880 UP000008820 UP000075903 UP000076407 UP000075884 UP000092462 UP000075920 UP000092445 UP000075840 UP000075902 UP000007062 UP000092461 UP000037069 UP000001292 UP000092444 UP000078200 UP000092443 UP000092460 UP000007819 UP000091820 UP000002282 UP000001070 UP000007798 UP000268350 UP000095300 UP000008792 UP000192221 UP000092553 UP000008744 UP000001819 UP000008711 UP000000304 UP000000803 UP000009192 UP000007801 UP000076408 UP000075901
UP000030742 UP000019118 UP000027135 UP000007266 UP000235965 UP000009046 UP000069940 UP000249989 UP000075880 UP000008820 UP000075903 UP000076407 UP000075884 UP000092462 UP000075920 UP000092445 UP000075840 UP000075902 UP000007062 UP000092461 UP000037069 UP000001292 UP000092444 UP000078200 UP000092443 UP000092460 UP000007819 UP000091820 UP000002282 UP000001070 UP000007798 UP000268350 UP000095300 UP000008792 UP000192221 UP000092553 UP000008744 UP000001819 UP000008711 UP000000304 UP000000803 UP000009192 UP000007801 UP000076408 UP000075901
Interpro
SUPFAM
SSF82895
SSF82895
Gene 3D
ProteinModelPortal
H9IXI6
A0A437ARJ2
A0A194QEY4
A0A2W1BSJ3
A0A194QW27
A0A2A4IYQ3
+ More
A0A212F8C6 U4UKR6 N6SSR5 A0A067RCW6 D6WRW7 A0A2J7R4U0 A0A1Y1LAA0 A0A1Y1LDZ5 A0A1B6EY69 A0A2J7R4T8 E0VAR6 A0A1S4FP70 A0A182H310 A0A182JFI6 Q16TY3 A0A2Y9D2Y1 A0A182XEH7 A0A182N1Y1 A0A1B0D865 A0A182WM27 A0A1A9ZX31 A0A182HQM0 A0A182UJG2 Q7Q7Y1 A0A1B0GKN0 A0A0A9XI82 A0A0A9XN21 A0A0L0CKU5 A0A0A9XCR4 B4I8B2 Q8T3M5 A0A2S2QNX1 A0A034V3Q9 A0A2S2P9A4 W8APV3 A0A1B0EUB2 A0A2S2RB68 A0A1A9VA32 A0A1A9XAQ9 A0A1B0BN75 J9JTN6 A0A1A9WYG6 B4P5T4 B4J412 A0A2S2PIE9 A0A336KUH1 B4MQA0 A0A3B0IY20 A0A0K8VW49 A0A336MQA6 A0A2H8TN60 A0A1I8PMI4 A0A0A1X1I4 B4LIV2 A0A0Q9W4L5 A0A1W4VQL9 Q8T458 A0A0M3QVE7 B4GAH9 A0A0K8U7W2 Q291N2 B3NNW2 A0A0J9RJ86 B4QHN1 Q9W1Z6 B4KPC0 A0A3B0JHX5 A0A3B0IYE1 B3MEJ2 A0A182YR46 A0A182S8R9
A0A212F8C6 U4UKR6 N6SSR5 A0A067RCW6 D6WRW7 A0A2J7R4U0 A0A1Y1LAA0 A0A1Y1LDZ5 A0A1B6EY69 A0A2J7R4T8 E0VAR6 A0A1S4FP70 A0A182H310 A0A182JFI6 Q16TY3 A0A2Y9D2Y1 A0A182XEH7 A0A182N1Y1 A0A1B0D865 A0A182WM27 A0A1A9ZX31 A0A182HQM0 A0A182UJG2 Q7Q7Y1 A0A1B0GKN0 A0A0A9XI82 A0A0A9XN21 A0A0L0CKU5 A0A0A9XCR4 B4I8B2 Q8T3M5 A0A2S2QNX1 A0A034V3Q9 A0A2S2P9A4 W8APV3 A0A1B0EUB2 A0A2S2RB68 A0A1A9VA32 A0A1A9XAQ9 A0A1B0BN75 J9JTN6 A0A1A9WYG6 B4P5T4 B4J412 A0A2S2PIE9 A0A336KUH1 B4MQA0 A0A3B0IY20 A0A0K8VW49 A0A336MQA6 A0A2H8TN60 A0A1I8PMI4 A0A0A1X1I4 B4LIV2 A0A0Q9W4L5 A0A1W4VQL9 Q8T458 A0A0M3QVE7 B4GAH9 A0A0K8U7W2 Q291N2 B3NNW2 A0A0J9RJ86 B4QHN1 Q9W1Z6 B4KPC0 A0A3B0JHX5 A0A3B0IYE1 B3MEJ2 A0A182YR46 A0A182S8R9
Ontologies
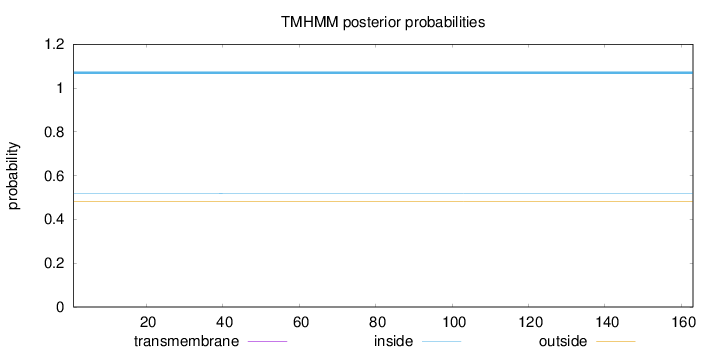
Topology
Length:
163
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00235
Exp number, first 60 AAs:
0.00161
Total prob of N-in:
0.51918
inside
1 - 163
Population Genetic Test Statistics
Pi
168.221563
Theta
75.816135
Tajima's D
0.177592
CLR
0
CSRT
0.421128943552822
Interpretation
Uncertain
