Gene
KWMTBOMO11343
Pre Gene Modal
BGIBMGA001850
Annotation
PREDICTED:_lipid_phosphate_phosphohydrolase_2-like_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.764
Sequence
CDS
ATGGTGGCACTCTGCCGTCGGCTCGGCTCTGATCCGGTGCAGGAACTCACCGAAGCAACCATGCCCATGCTTGCTCTCATCGGTTTGTTCGAGTTGGGCATCATACCGAACAGAAAAGCGGGTTTCTTCTGTAACGATGCTGCTATATCCTTCGACTACACAGGAGAGACGGTGACCAGCCTCATCCTTATGACAACTATATTCATAATACCTATCGGAGTTTTCATGATAACAGAATATATATTCACACCTAACGAAATACCACTGTCGGATAGAACGATGAGGGCCTTCCTGAAGAGCTCGTGGCTTTTCAAAACGTACTTATACGGCTTCATGATGAACCTTTGCATCGTGGAAGTTATGAAGGGTATCGTTGGTAATCCAAGACCTGTGTTCTTCGCGCTGTGCCAACCAGACACTGCTAAGACGTGTAACGGCACGGATTTCGTGAGTACTTTCGAGTGCACGTCATCGTACTCCCGCTGGTACCGGATGGATTCGTATCGGAGTTTTCCGTCGGGGCACACATCGTTGTCCGTATACTGCGGGTTTTTCCTGGCGTGGTACCTTCAAAAGCGGGCCTTCAACTGGTCCTACCGCTCCGAGCTGGTGGTCCCGGTGGTCCAGCTCCTCTGCGTGTCGTACTCCGTGCTGTGCTCGCTCACCAGGATCACGGACCGCATGCACCACTGGTGGGACGTCCTGATCGGATCCGCCATTGGACTGATCACTTTGTTGTACACCGTGATATTTTTAAACGACAACTTCTCGTACTTCAACACGAGCGACTCAGGGGAGGAGGGGGGTACCGCGCCCACGAGAACAGTATTATTCGATGAAAGAAGAGATAGCTAA
Protein
MVALCRRLGSDPVQELTEATMPMLALIGLFELGIIPNRKAGFFCNDAAISFDYTGETVTSLILMTTIFIIPIGVFMITEYIFTPNEIPLSDRTMRAFLKSSWLFKTYLYGFMMNLCIVEVMKGIVGNPRPVFFALCQPDTAKTCNGTDFVSTFECTSSYSRWYRMDSYRSFPSGHTSLSVYCGFFLAWYLQKRAFNWSYRSELVVPVVQLLCVSYSVLCSLTRITDRMHHWWDVLIGSAIGLITLLYTVIFLNDNFSYFNTSDSGEEGGTAPTRTVLFDERRDS
Summary
Uniprot
H9IX67
A0A2H1V7G8
A0A2W1BZB5
A0A2A4J9V6
A0A2A4JB39
A0A194R0Z6
+ More
A0A194QKX8 A0A437BRC0 A0A212FAY8 A0A2H1W6D3 A0A212FE60 A0A194QFB0 A0A2W1BWN5 A0A2H1VNK2 A0A2A4JDK4 A0A3S2NPQ9 A0A0L7LSS2 A0A0N0PF37 A0A1B0CLC2 A0A2A4J2W7 A0A194QEX9 A0A2H1WD33 A0A2H1VPN2 A0A2W1B3A1 A0A1L8DY82 A0A1W4WRV3 A0A1L8DYE2 A0A1W4WFP8 A0A1W4WRJ7 A0A1L8DY42 A0A1L8DY94 A0A1L8DY28 A0A1L8DYB3 A0A1L8DY72 A0A0K8TPI1 A0A1L8DY32 A0A1L8DY37 A0A1L8DY38 A0A151X268 A0A1L8DYD3 A0A026W573 A0A3L8E0C8 A0A182YM84 A0A1J1HRS7 A0A1J1HW37 A0A194QW33 E2AJW7 A0A1B0D8B6 A0A2H8TZ89 A0A0G3YJ71 A0A2P8ZBP0 A0A154PPE1 A0A195E411 A0A182VBL5 A0A182PJF7 J9K159 A0A182IBH0 A0A1S4GZL6 A0A182KN29 A0A195FIV6 A0A0P6DB88 A0A0P5G2Z8 A0A195AVC0 K7IRG6 A0A0L7R3F4 A0A1Y9IUJ3 A0A0P5LR52 A0A0P5GQC3 A0A182WFD5 A0A0P4VTC9 A0A0N8DY65 A0A310SQX9 A0A158P2M9 F4W6H5 A0A0P5XS63 A0A164S6G0 A0A182Q0M4 A0A182JJE1 J3JWD3 A0A0P6AX81 U4UMF1 N6TYZ0 A0A2A3E788 A0A088A7K3 A0A146M6G7 A0A0A9YLC8 A0A2M4BU51 A0A146M951 A0A084VJ71 A0A1B6G844 A0A2M4CPZ9 A0A2M4CQ63 A0A067R3Z5 A0A2M4CQ12 E9G7S0 A0A1Y1MRF5 A0A182NGS4 A0A1Y1MN55 A0A182F385
A0A194QKX8 A0A437BRC0 A0A212FAY8 A0A2H1W6D3 A0A212FE60 A0A194QFB0 A0A2W1BWN5 A0A2H1VNK2 A0A2A4JDK4 A0A3S2NPQ9 A0A0L7LSS2 A0A0N0PF37 A0A1B0CLC2 A0A2A4J2W7 A0A194QEX9 A0A2H1WD33 A0A2H1VPN2 A0A2W1B3A1 A0A1L8DY82 A0A1W4WRV3 A0A1L8DYE2 A0A1W4WFP8 A0A1W4WRJ7 A0A1L8DY42 A0A1L8DY94 A0A1L8DY28 A0A1L8DYB3 A0A1L8DY72 A0A0K8TPI1 A0A1L8DY32 A0A1L8DY37 A0A1L8DY38 A0A151X268 A0A1L8DYD3 A0A026W573 A0A3L8E0C8 A0A182YM84 A0A1J1HRS7 A0A1J1HW37 A0A194QW33 E2AJW7 A0A1B0D8B6 A0A2H8TZ89 A0A0G3YJ71 A0A2P8ZBP0 A0A154PPE1 A0A195E411 A0A182VBL5 A0A182PJF7 J9K159 A0A182IBH0 A0A1S4GZL6 A0A182KN29 A0A195FIV6 A0A0P6DB88 A0A0P5G2Z8 A0A195AVC0 K7IRG6 A0A0L7R3F4 A0A1Y9IUJ3 A0A0P5LR52 A0A0P5GQC3 A0A182WFD5 A0A0P4VTC9 A0A0N8DY65 A0A310SQX9 A0A158P2M9 F4W6H5 A0A0P5XS63 A0A164S6G0 A0A182Q0M4 A0A182JJE1 J3JWD3 A0A0P6AX81 U4UMF1 N6TYZ0 A0A2A3E788 A0A088A7K3 A0A146M6G7 A0A0A9YLC8 A0A2M4BU51 A0A146M951 A0A084VJ71 A0A1B6G844 A0A2M4CPZ9 A0A2M4CQ63 A0A067R3Z5 A0A2M4CQ12 E9G7S0 A0A1Y1MRF5 A0A182NGS4 A0A1Y1MN55 A0A182F385
Pubmed
EMBL
BABH01018212
BABH01018213
ODYU01001069
SOQ36793.1
KZ149913
PZC78036.1
+ More
NWSH01002206 PCG68907.1 PCG68908.1 KQ461073 KPJ09516.1 KQ459053 KPJ04101.1 RSAL01000016 RVE53008.1 AGBW02009413 OWR50889.1 ODYU01006616 SOQ48618.1 AGBW02008985 OWR51977.1 KPJ04099.1 PZC78034.1 ODYU01003520 SOQ42415.1 NWSH01001933 PCG69634.1 RSAL01000611 RVE41309.1 JTDY01000163 KOB78502.1 LADI01014042 KPJ20781.1 AJWK01017073 NWSH01003358 PCG66487.1 KPJ04098.1 ODYU01007532 SOQ50384.1 SOQ42412.1 KZ150379 PZC71102.1 GFDF01002715 JAV11369.1 GFDF01002648 JAV11436.1 GFDF01002716 JAV11368.1 GFDF01002647 JAV11437.1 GFDF01002717 JAV11367.1 GFDF01002646 JAV11438.1 GFDF01002720 JAV11364.1 GDAI01001339 JAI16264.1 GFDF01002714 JAV11370.1 GFDF01002707 JAV11377.1 GFDF01002721 JAV11363.1 KQ982580 KYQ54478.1 GFDF01002650 JAV11434.1 KK107474 EZA50179.1 QOIP01000002 RLU26107.1 CVRI01000020 CRK90725.1 CRK90726.1 KPJ09519.1 GL440100 EFN66283.1 AJVK01004036 GFXV01007771 MBW19576.1 KM507106 AKM28430.1 PYGN01000114 PSN53890.1 KQ435012 KZC13765.1 KQ979657 KYN19920.1 ABLF02039852 ABLF02039853 APCN01000689 AAAB01008944 KQ981523 KYN40333.1 GDIQ01080349 JAN14388.1 GDIQ01248100 JAK03625.1 KQ976731 KYM76183.1 KQ414663 KOC65407.1 GDIQ01190773 JAK60952.1 GDIQ01244157 JAK07568.1 GDKW01003444 JAI53151.1 GDIQ01080348 JAN14389.1 KQ761467 OAD57510.1 ADTU01007015 ADTU01007016 ADTU01007017 ADTU01007018 ADTU01007019 ADTU01007020 ADTU01007021 ADTU01007022 ADTU01007023 GL887707 EGI70310.1 GDIP01068101 JAM35614.1 LRGB01002076 KZS09305.1 AXCN02002201 BT127551 AEE62513.1 GDIP01023027 JAM80688.1 KB632367 ERL93668.1 APGK01045770 APGK01045771 KB741039 ENN74515.1 KZ288347 PBC27575.1 GDHC01004283 JAQ14346.1 GBHO01033783 GBHO01013264 JAG09821.1 JAG30340.1 GGFJ01007456 MBW56597.1 GDHC01003057 JAQ15572.1 ATLV01013474 KE524860 KFB38015.1 GECZ01029757 GECZ01012517 GECZ01011154 GECZ01005000 JAS40012.1 JAS57252.1 JAS58615.1 JAS64769.1 GGFL01003244 MBW67422.1 GGFL01003241 MBW67419.1 KK852718 KDR17784.1 GGFL01003242 MBW67420.1 GL732534 EFX84528.1 GEZM01026250 GEZM01026249 JAV87095.1 GEZM01026252 GEZM01026251 JAV87092.1
NWSH01002206 PCG68907.1 PCG68908.1 KQ461073 KPJ09516.1 KQ459053 KPJ04101.1 RSAL01000016 RVE53008.1 AGBW02009413 OWR50889.1 ODYU01006616 SOQ48618.1 AGBW02008985 OWR51977.1 KPJ04099.1 PZC78034.1 ODYU01003520 SOQ42415.1 NWSH01001933 PCG69634.1 RSAL01000611 RVE41309.1 JTDY01000163 KOB78502.1 LADI01014042 KPJ20781.1 AJWK01017073 NWSH01003358 PCG66487.1 KPJ04098.1 ODYU01007532 SOQ50384.1 SOQ42412.1 KZ150379 PZC71102.1 GFDF01002715 JAV11369.1 GFDF01002648 JAV11436.1 GFDF01002716 JAV11368.1 GFDF01002647 JAV11437.1 GFDF01002717 JAV11367.1 GFDF01002646 JAV11438.1 GFDF01002720 JAV11364.1 GDAI01001339 JAI16264.1 GFDF01002714 JAV11370.1 GFDF01002707 JAV11377.1 GFDF01002721 JAV11363.1 KQ982580 KYQ54478.1 GFDF01002650 JAV11434.1 KK107474 EZA50179.1 QOIP01000002 RLU26107.1 CVRI01000020 CRK90725.1 CRK90726.1 KPJ09519.1 GL440100 EFN66283.1 AJVK01004036 GFXV01007771 MBW19576.1 KM507106 AKM28430.1 PYGN01000114 PSN53890.1 KQ435012 KZC13765.1 KQ979657 KYN19920.1 ABLF02039852 ABLF02039853 APCN01000689 AAAB01008944 KQ981523 KYN40333.1 GDIQ01080349 JAN14388.1 GDIQ01248100 JAK03625.1 KQ976731 KYM76183.1 KQ414663 KOC65407.1 GDIQ01190773 JAK60952.1 GDIQ01244157 JAK07568.1 GDKW01003444 JAI53151.1 GDIQ01080348 JAN14389.1 KQ761467 OAD57510.1 ADTU01007015 ADTU01007016 ADTU01007017 ADTU01007018 ADTU01007019 ADTU01007020 ADTU01007021 ADTU01007022 ADTU01007023 GL887707 EGI70310.1 GDIP01068101 JAM35614.1 LRGB01002076 KZS09305.1 AXCN02002201 BT127551 AEE62513.1 GDIP01023027 JAM80688.1 KB632367 ERL93668.1 APGK01045770 APGK01045771 KB741039 ENN74515.1 KZ288347 PBC27575.1 GDHC01004283 JAQ14346.1 GBHO01033783 GBHO01013264 JAG09821.1 JAG30340.1 GGFJ01007456 MBW56597.1 GDHC01003057 JAQ15572.1 ATLV01013474 KE524860 KFB38015.1 GECZ01029757 GECZ01012517 GECZ01011154 GECZ01005000 JAS40012.1 JAS57252.1 JAS58615.1 JAS64769.1 GGFL01003244 MBW67422.1 GGFL01003241 MBW67419.1 KK852718 KDR17784.1 GGFL01003242 MBW67420.1 GL732534 EFX84528.1 GEZM01026250 GEZM01026249 JAV87095.1 GEZM01026252 GEZM01026251 JAV87092.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000283053
UP000007151
+ More
UP000037510 UP000092461 UP000192223 UP000075809 UP000053097 UP000279307 UP000076408 UP000183832 UP000000311 UP000092462 UP000245037 UP000076502 UP000078492 UP000075903 UP000075885 UP000007819 UP000075840 UP000075882 UP000078541 UP000078540 UP000002358 UP000053825 UP000075920 UP000005205 UP000007755 UP000076858 UP000075886 UP000075880 UP000030742 UP000019118 UP000242457 UP000005203 UP000030765 UP000027135 UP000000305 UP000075884 UP000069272
UP000037510 UP000092461 UP000192223 UP000075809 UP000053097 UP000279307 UP000076408 UP000183832 UP000000311 UP000092462 UP000245037 UP000076502 UP000078492 UP000075903 UP000075885 UP000007819 UP000075840 UP000075882 UP000078541 UP000078540 UP000002358 UP000053825 UP000075920 UP000005205 UP000007755 UP000076858 UP000075886 UP000075880 UP000030742 UP000019118 UP000242457 UP000005203 UP000030765 UP000027135 UP000000305 UP000075884 UP000069272
PRIDE
Pfam
PF01569 PAP2
SUPFAM
SSF48317
SSF48317
ProteinModelPortal
H9IX67
A0A2H1V7G8
A0A2W1BZB5
A0A2A4J9V6
A0A2A4JB39
A0A194R0Z6
+ More
A0A194QKX8 A0A437BRC0 A0A212FAY8 A0A2H1W6D3 A0A212FE60 A0A194QFB0 A0A2W1BWN5 A0A2H1VNK2 A0A2A4JDK4 A0A3S2NPQ9 A0A0L7LSS2 A0A0N0PF37 A0A1B0CLC2 A0A2A4J2W7 A0A194QEX9 A0A2H1WD33 A0A2H1VPN2 A0A2W1B3A1 A0A1L8DY82 A0A1W4WRV3 A0A1L8DYE2 A0A1W4WFP8 A0A1W4WRJ7 A0A1L8DY42 A0A1L8DY94 A0A1L8DY28 A0A1L8DYB3 A0A1L8DY72 A0A0K8TPI1 A0A1L8DY32 A0A1L8DY37 A0A1L8DY38 A0A151X268 A0A1L8DYD3 A0A026W573 A0A3L8E0C8 A0A182YM84 A0A1J1HRS7 A0A1J1HW37 A0A194QW33 E2AJW7 A0A1B0D8B6 A0A2H8TZ89 A0A0G3YJ71 A0A2P8ZBP0 A0A154PPE1 A0A195E411 A0A182VBL5 A0A182PJF7 J9K159 A0A182IBH0 A0A1S4GZL6 A0A182KN29 A0A195FIV6 A0A0P6DB88 A0A0P5G2Z8 A0A195AVC0 K7IRG6 A0A0L7R3F4 A0A1Y9IUJ3 A0A0P5LR52 A0A0P5GQC3 A0A182WFD5 A0A0P4VTC9 A0A0N8DY65 A0A310SQX9 A0A158P2M9 F4W6H5 A0A0P5XS63 A0A164S6G0 A0A182Q0M4 A0A182JJE1 J3JWD3 A0A0P6AX81 U4UMF1 N6TYZ0 A0A2A3E788 A0A088A7K3 A0A146M6G7 A0A0A9YLC8 A0A2M4BU51 A0A146M951 A0A084VJ71 A0A1B6G844 A0A2M4CPZ9 A0A2M4CQ63 A0A067R3Z5 A0A2M4CQ12 E9G7S0 A0A1Y1MRF5 A0A182NGS4 A0A1Y1MN55 A0A182F385
A0A194QKX8 A0A437BRC0 A0A212FAY8 A0A2H1W6D3 A0A212FE60 A0A194QFB0 A0A2W1BWN5 A0A2H1VNK2 A0A2A4JDK4 A0A3S2NPQ9 A0A0L7LSS2 A0A0N0PF37 A0A1B0CLC2 A0A2A4J2W7 A0A194QEX9 A0A2H1WD33 A0A2H1VPN2 A0A2W1B3A1 A0A1L8DY82 A0A1W4WRV3 A0A1L8DYE2 A0A1W4WFP8 A0A1W4WRJ7 A0A1L8DY42 A0A1L8DY94 A0A1L8DY28 A0A1L8DYB3 A0A1L8DY72 A0A0K8TPI1 A0A1L8DY32 A0A1L8DY37 A0A1L8DY38 A0A151X268 A0A1L8DYD3 A0A026W573 A0A3L8E0C8 A0A182YM84 A0A1J1HRS7 A0A1J1HW37 A0A194QW33 E2AJW7 A0A1B0D8B6 A0A2H8TZ89 A0A0G3YJ71 A0A2P8ZBP0 A0A154PPE1 A0A195E411 A0A182VBL5 A0A182PJF7 J9K159 A0A182IBH0 A0A1S4GZL6 A0A182KN29 A0A195FIV6 A0A0P6DB88 A0A0P5G2Z8 A0A195AVC0 K7IRG6 A0A0L7R3F4 A0A1Y9IUJ3 A0A0P5LR52 A0A0P5GQC3 A0A182WFD5 A0A0P4VTC9 A0A0N8DY65 A0A310SQX9 A0A158P2M9 F4W6H5 A0A0P5XS63 A0A164S6G0 A0A182Q0M4 A0A182JJE1 J3JWD3 A0A0P6AX81 U4UMF1 N6TYZ0 A0A2A3E788 A0A088A7K3 A0A146M6G7 A0A0A9YLC8 A0A2M4BU51 A0A146M951 A0A084VJ71 A0A1B6G844 A0A2M4CPZ9 A0A2M4CQ63 A0A067R3Z5 A0A2M4CQ12 E9G7S0 A0A1Y1MRF5 A0A182NGS4 A0A1Y1MN55 A0A182F385
Ontologies
PATHWAY
00561
Glycerolipid metabolism - Bombyx mori (domestic silkworm)
00564 Glycerophospholipid metabolism - Bombyx mori (domestic silkworm)
00565 Ether lipid metabolism - Bombyx mori (domestic silkworm)
00600 Sphingolipid metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00564 Glycerophospholipid metabolism - Bombyx mori (domestic silkworm)
00565 Ether lipid metabolism - Bombyx mori (domestic silkworm)
00600 Sphingolipid metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
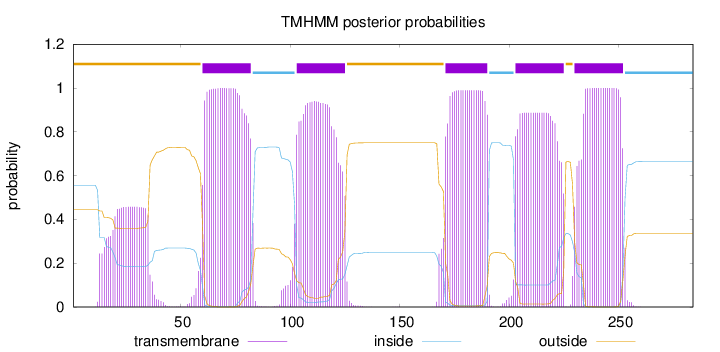
Topology
Length:
284
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
114.1379
Exp number, first 60 AAs:
10.90832
Total prob of N-in:
0.55581
POSSIBLE N-term signal
sequence
outside
1 - 59
TMhelix
60 - 82
inside
83 - 102
TMhelix
103 - 125
outside
126 - 170
TMhelix
171 - 190
inside
191 - 202
TMhelix
203 - 225
outside
226 - 229
TMhelix
230 - 252
inside
253 - 284
Population Genetic Test Statistics
Pi
219.281203
Theta
166.846566
Tajima's D
1.278848
CLR
0.389905
CSRT
0.732813359332033
Interpretation
Uncertain
