Gene
KWMTBOMO11338
Pre Gene Modal
BGIBMGA001974
Annotation
ionotropic_receptor_[Ostrinia_furnacalis]
Full name
Chloride channel protein
Location in the cell
Mitochondrial Reliability : 1.279
Sequence
CDS
ATGAAATTTGTTACTTTTCTTATGATAACGATTTGGGCCCGCGTGGTGCTCACAAAGGATGCGAACTTGGTGGATTTTCTTAAAACTTACGTGGATAATGAGAGAAAACCTACAATGGTTGTGTTGACGGGCCTTTGTTGGAAAAAAGACAGAGTGCGTAATCTAGTAGCTGATCTGTCAAAGCTTGGAATTAGGACATCGAAGCCTCCGTCTATAGATATACAATCCAAATACCACGTCGTCATGCACAATACATTGTTCCTGAACGATTTGCATTGTCCCGAAACGGCAGAGGTCATTGAAACTGCTATTCTACATAATCTATTTCGGTATCCTCTACGGTGGCTTTTGCTGAAAAACATGTCCGAAAACTCTTCTGTCATACTTGAAGACATACCCGCGTTTGCAGACAGCGATTTAGTCTTAGCAGAAATAACAGAAGAATTAATTTTACTTACTGAATTGCACAAACCGAATGAGAACGGCGCGTTGATATCAAAAGATAAAGGTTACTACAACGGAACTATGGTAGACGTTAGACCGCATCGAGAACTCTTCAAACGGCGCAGAGACGTGATGGGTCGTCCATTGACAATGGCGAATGTTATACAAGACAGCAACAACACTCGTTATCATCTGCCTAGGGAGGATGCCTTGGAACTCCAGTACGACGTTATTCCTAAAATCTGCTGGATGACCGCGAAGCTAGCGTTCCAAATGTTAAATGCCACTCCTCGGTACACCTTCAGCTATCGTTGGGGGTATAAAGTGAACGGGCAATGGTCGGGGATGATCAACGATTTGCACACCAGTAAAGCCGATTTGGGTAAGACTTAA
Protein
MKFVTFLMITIWARVVLTKDANLVDFLKTYVDNERKPTMVVLTGLCWKKDRVRNLVADLSKLGIRTSKPPSIDIQSKYHVVMHNTLFLNDLHCPETAEVIETAILHNLFRYPLRWLLLKNMSENSSVILEDIPAFADSDLVLAEITEELILLTELHKPNENGALISKDKGYYNGTMVDVRPHRELFKRRRDVMGRPLTMANVIQDSNNTRYHLPREDALELQYDVIPKICWMTAKLAFQMLNATPRYTFSYRWGYKVNGQWSGMINDLHTSKADLGKT
Summary
Similarity
Belongs to the chloride channel (TC 2.A.49) family.
Uniprot
A0A2W1B5Q5
A0A0F7QEH8
A0A0S1TK20
E5FIA9
A0A2H1VKZ8
A0A2W1BCP8
+ More
A0A140G9H4 A0A2K8GL81 A0A0F7QED4 A0A3S2LDH7 A0A2A4J494 A0A076E9E5 A0A212FC55 A0A2A4JCF7 A0A194QGN2 A0A345BF19 A0A0V0J280 A0A1Q1PP74 A0A0K8TUX6 H9A5R5 A0A223HDB0 A0A194QX34 A0A223HCY4 A0A2A4JCR5 A0A2W1BDU9 A0A140G9H3 A0A2K8GL77 A0A3Q8HEA9 A0A345BF26 A0A345BF17 A0A1B3B728 A0A386H9K5 A0A223HCY5 A0A437B2L0 A0A146JYY8 A0A0F7QIG2 A0A223HCX7 A0A0S1TQ33 A0A2H1VST4 A0A1W5LAS2 A0A0K8TUD4 A0A2D0WKS5 A0A0L7LLS6 A0A1W6L166 A0A0U3C6J1 A0A2P1JHG6 A0A1V1WBQ2 A0A2K8GL75 A0A223HDB5 A0A0S3J2Z8 A0A1Q1PP97 A0A0K8TUC6 A0A0B5A3J3 A0A310SDV6 A0A2D0WKS7 A0A0F7QIG0 A0A2A4IZW1 A0A1Q1PP98 A0A0C5DMP4 A0A0S3J2S5 A0A345BF14 E5FIB0 A0A212FEZ5 A0A223HDB2 A0A2H4ZBE9 H9A5R2 E2AM42 A0A232FN58 A0A067R2S3 A0A3L8E073 A0A0C9PTU2 A0A1S4GPY3 H9A5R6 A0A067QZN3 A0A182XCS1 A0A1B3P5G8 A0A1J0KL22 A0A2H1VRQ6 A0A182GF22 K7J111 A0A1B3P5F9
A0A140G9H4 A0A2K8GL81 A0A0F7QED4 A0A3S2LDH7 A0A2A4J494 A0A076E9E5 A0A212FC55 A0A2A4JCF7 A0A194QGN2 A0A345BF19 A0A0V0J280 A0A1Q1PP74 A0A0K8TUX6 H9A5R5 A0A223HDB0 A0A194QX34 A0A223HCY4 A0A2A4JCR5 A0A2W1BDU9 A0A140G9H3 A0A2K8GL77 A0A3Q8HEA9 A0A345BF26 A0A345BF17 A0A1B3B728 A0A386H9K5 A0A223HCY5 A0A437B2L0 A0A146JYY8 A0A0F7QIG2 A0A223HCX7 A0A0S1TQ33 A0A2H1VST4 A0A1W5LAS2 A0A0K8TUD4 A0A2D0WKS5 A0A0L7LLS6 A0A1W6L166 A0A0U3C6J1 A0A2P1JHG6 A0A1V1WBQ2 A0A2K8GL75 A0A223HDB5 A0A0S3J2Z8 A0A1Q1PP97 A0A0K8TUC6 A0A0B5A3J3 A0A310SDV6 A0A2D0WKS7 A0A0F7QIG0 A0A2A4IZW1 A0A1Q1PP98 A0A0C5DMP4 A0A0S3J2S5 A0A345BF14 E5FIB0 A0A212FEZ5 A0A223HDB2 A0A2H4ZBE9 H9A5R2 E2AM42 A0A232FN58 A0A067R2S3 A0A3L8E073 A0A0C9PTU2 A0A1S4GPY3 H9A5R6 A0A067QZN3 A0A182XCS1 A0A1B3P5G8 A0A1J0KL22 A0A2H1VRQ6 A0A182GF22 K7J111 A0A1B3P5F9
Pubmed
EMBL
KZ150379
PZC71098.1
LC017791
BAR64805.1
KR912012
ALM24939.1
+ More
HM562973 ADR64684.1 ODYU01003127 SOQ41495.1 PZC71097.1 KU702626 AMM70653.1 KY225533 ARO70545.1 LC017792 BAR64806.1 RSAL01000205 RVE44372.1 NWSH01003358 PCG66488.1 KF487730 AII01128.1 AGBW02009222 OWR51314.1 NWSH01001928 PCG69641.1 KQ459053 KPJ04095.1 MG820672 AXF48843.1 GDKB01000021 JAP38475.1 KY325454 AQM73615.1 GCVX01000137 JAI18093.1 JN836715 AFC91755.2 KY283703 AST36362.1 KQ461073 KPJ09520.1 KY283574 AST36234.1 PCG69639.1 PZC71096.1 KU702625 AMM70652.1 KY225534 ARO70546.1 MF625646 AXY83471.1 MG820679 AXF48850.1 MG820670 AXF48841.1 KT588085 AOE47995.1 MG546648 AYD42271.1 KY283575 AST36235.1 RSAL01000189 RVE44731.1 GEDO01000005 JAP88621.1 LC017794 BAR64808.1 KY283576 AST36236.1 KR912013 ALM24940.1 ODYU01004051 SOQ43532.1 KT991368 ANA75034.2 GCVX01000138 JAI18092.1 KY019179 ARB05668.1 JTDY01000633 KOB76402.1 KX609444 KB465935 ARN17851.1 RLZ02208.1 KP975110 ALT31629.1 MG766213 AVN97883.1 GENK01000124 JAV45789.1 KY225535 ARO70547.1 KY283704 AST36363.1 KT381535 ALR72541.1 KY325455 AQM73616.1 GCVX01000136 JAI18094.1 KJ542755 AJD81639.1 GGOB01000048 MCH29444.1 KY019178 ARB05667.1 LC017789 BAR64803.1 NWSH01004502 PCG65036.1 KY325456 AQM73617.1 KP296776 AJO62240.1 KT381531 ALR72537.1 MG820667 AXF48838.1 HM562974 ADR64685.1 AGBW02008878 OWR52312.1 KY283705 AST36364.1 MF975509 AUF73085.1 JN836712 AFC91752.1 GL440704 EFN65500.1 NNAY01000014 OXU31959.1 KK852952 KDR13328.1 QOIP01000002 RLU26111.1 GBYB01004753 JAG74520.1 AAAB01008960 JN836716 AFC91756.2 KK852854 KDR14986.1 KX655894 AOG12843.1 KX298831 APC94348.1 SOQ43533.1 JXUM01059078 JXUM01059079 JXUM01059080 JXUM01059081 JXUM01059082 KQ562034 KXJ76850.1 KX655899 AOG12848.1
HM562973 ADR64684.1 ODYU01003127 SOQ41495.1 PZC71097.1 KU702626 AMM70653.1 KY225533 ARO70545.1 LC017792 BAR64806.1 RSAL01000205 RVE44372.1 NWSH01003358 PCG66488.1 KF487730 AII01128.1 AGBW02009222 OWR51314.1 NWSH01001928 PCG69641.1 KQ459053 KPJ04095.1 MG820672 AXF48843.1 GDKB01000021 JAP38475.1 KY325454 AQM73615.1 GCVX01000137 JAI18093.1 JN836715 AFC91755.2 KY283703 AST36362.1 KQ461073 KPJ09520.1 KY283574 AST36234.1 PCG69639.1 PZC71096.1 KU702625 AMM70652.1 KY225534 ARO70546.1 MF625646 AXY83471.1 MG820679 AXF48850.1 MG820670 AXF48841.1 KT588085 AOE47995.1 MG546648 AYD42271.1 KY283575 AST36235.1 RSAL01000189 RVE44731.1 GEDO01000005 JAP88621.1 LC017794 BAR64808.1 KY283576 AST36236.1 KR912013 ALM24940.1 ODYU01004051 SOQ43532.1 KT991368 ANA75034.2 GCVX01000138 JAI18092.1 KY019179 ARB05668.1 JTDY01000633 KOB76402.1 KX609444 KB465935 ARN17851.1 RLZ02208.1 KP975110 ALT31629.1 MG766213 AVN97883.1 GENK01000124 JAV45789.1 KY225535 ARO70547.1 KY283704 AST36363.1 KT381535 ALR72541.1 KY325455 AQM73616.1 GCVX01000136 JAI18094.1 KJ542755 AJD81639.1 GGOB01000048 MCH29444.1 KY019178 ARB05667.1 LC017789 BAR64803.1 NWSH01004502 PCG65036.1 KY325456 AQM73617.1 KP296776 AJO62240.1 KT381531 ALR72537.1 MG820667 AXF48838.1 HM562974 ADR64685.1 AGBW02008878 OWR52312.1 KY283705 AST36364.1 MF975509 AUF73085.1 JN836712 AFC91752.1 GL440704 EFN65500.1 NNAY01000014 OXU31959.1 KK852952 KDR13328.1 QOIP01000002 RLU26111.1 GBYB01004753 JAG74520.1 AAAB01008960 JN836716 AFC91756.2 KK852854 KDR14986.1 KX655894 AOG12843.1 KX298831 APC94348.1 SOQ43533.1 JXUM01059078 JXUM01059079 JXUM01059080 JXUM01059081 JXUM01059082 KQ562034 KXJ76850.1 KX655899 AOG12848.1
Proteomes
Pfam
Interpro
IPR001320
Iontro_rcpt
+ More
IPR013087 Znf_C2H2_type
IPR002320 Thr-tRNA-ligase_IIa
IPR002314 aa-tRNA-synt_IIb
IPR036621 Anticodon-bd_dom_sf
IPR012675 Beta-grasp_dom_sf
IPR012947 tRNA_SAD
IPR033728 ThrRS_core
IPR012676 TGS-like
IPR004154 Anticodon-bd
IPR018163 Thr/Ala-tRNA-synth_IIc_edit
IPR006195 aa-tRNA-synth_II
IPR004095 TGS
IPR019594 Glu/Gly-bd
IPR001638 Solute-binding_3/MltF_N
IPR000644 CBS_dom
IPR001807 Cl-channel_volt-gated
IPR014743 Cl-channel_core
IPR013087 Znf_C2H2_type
IPR002320 Thr-tRNA-ligase_IIa
IPR002314 aa-tRNA-synt_IIb
IPR036621 Anticodon-bd_dom_sf
IPR012675 Beta-grasp_dom_sf
IPR012947 tRNA_SAD
IPR033728 ThrRS_core
IPR012676 TGS-like
IPR004154 Anticodon-bd
IPR018163 Thr/Ala-tRNA-synth_IIc_edit
IPR006195 aa-tRNA-synth_II
IPR004095 TGS
IPR019594 Glu/Gly-bd
IPR001638 Solute-binding_3/MltF_N
IPR000644 CBS_dom
IPR001807 Cl-channel_volt-gated
IPR014743 Cl-channel_core
Gene 3D
ProteinModelPortal
A0A2W1B5Q5
A0A0F7QEH8
A0A0S1TK20
E5FIA9
A0A2H1VKZ8
A0A2W1BCP8
+ More
A0A140G9H4 A0A2K8GL81 A0A0F7QED4 A0A3S2LDH7 A0A2A4J494 A0A076E9E5 A0A212FC55 A0A2A4JCF7 A0A194QGN2 A0A345BF19 A0A0V0J280 A0A1Q1PP74 A0A0K8TUX6 H9A5R5 A0A223HDB0 A0A194QX34 A0A223HCY4 A0A2A4JCR5 A0A2W1BDU9 A0A140G9H3 A0A2K8GL77 A0A3Q8HEA9 A0A345BF26 A0A345BF17 A0A1B3B728 A0A386H9K5 A0A223HCY5 A0A437B2L0 A0A146JYY8 A0A0F7QIG2 A0A223HCX7 A0A0S1TQ33 A0A2H1VST4 A0A1W5LAS2 A0A0K8TUD4 A0A2D0WKS5 A0A0L7LLS6 A0A1W6L166 A0A0U3C6J1 A0A2P1JHG6 A0A1V1WBQ2 A0A2K8GL75 A0A223HDB5 A0A0S3J2Z8 A0A1Q1PP97 A0A0K8TUC6 A0A0B5A3J3 A0A310SDV6 A0A2D0WKS7 A0A0F7QIG0 A0A2A4IZW1 A0A1Q1PP98 A0A0C5DMP4 A0A0S3J2S5 A0A345BF14 E5FIB0 A0A212FEZ5 A0A223HDB2 A0A2H4ZBE9 H9A5R2 E2AM42 A0A232FN58 A0A067R2S3 A0A3L8E073 A0A0C9PTU2 A0A1S4GPY3 H9A5R6 A0A067QZN3 A0A182XCS1 A0A1B3P5G8 A0A1J0KL22 A0A2H1VRQ6 A0A182GF22 K7J111 A0A1B3P5F9
A0A140G9H4 A0A2K8GL81 A0A0F7QED4 A0A3S2LDH7 A0A2A4J494 A0A076E9E5 A0A212FC55 A0A2A4JCF7 A0A194QGN2 A0A345BF19 A0A0V0J280 A0A1Q1PP74 A0A0K8TUX6 H9A5R5 A0A223HDB0 A0A194QX34 A0A223HCY4 A0A2A4JCR5 A0A2W1BDU9 A0A140G9H3 A0A2K8GL77 A0A3Q8HEA9 A0A345BF26 A0A345BF17 A0A1B3B728 A0A386H9K5 A0A223HCY5 A0A437B2L0 A0A146JYY8 A0A0F7QIG2 A0A223HCX7 A0A0S1TQ33 A0A2H1VST4 A0A1W5LAS2 A0A0K8TUD4 A0A2D0WKS5 A0A0L7LLS6 A0A1W6L166 A0A0U3C6J1 A0A2P1JHG6 A0A1V1WBQ2 A0A2K8GL75 A0A223HDB5 A0A0S3J2Z8 A0A1Q1PP97 A0A0K8TUC6 A0A0B5A3J3 A0A310SDV6 A0A2D0WKS7 A0A0F7QIG0 A0A2A4IZW1 A0A1Q1PP98 A0A0C5DMP4 A0A0S3J2S5 A0A345BF14 E5FIB0 A0A212FEZ5 A0A223HDB2 A0A2H4ZBE9 H9A5R2 E2AM42 A0A232FN58 A0A067R2S3 A0A3L8E073 A0A0C9PTU2 A0A1S4GPY3 H9A5R6 A0A067QZN3 A0A182XCS1 A0A1B3P5G8 A0A1J0KL22 A0A2H1VRQ6 A0A182GF22 K7J111 A0A1B3P5F9
Ontologies
KEGG
GO
PANTHER
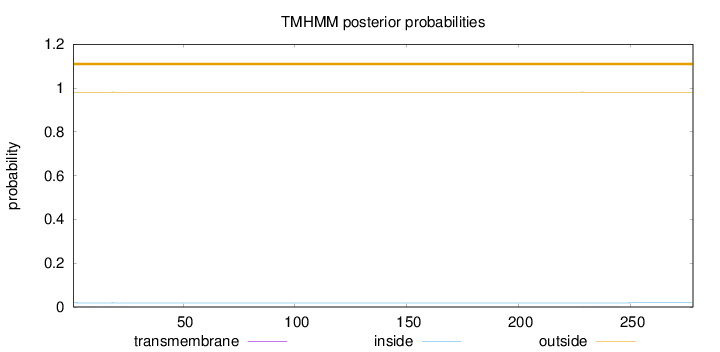
Topology
Subcellular location
Membrane
SignalP
Position: 1 - 18,
Likelihood: 0.957716

Length:
278
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01568
Exp number, first 60 AAs:
0.01146
Total prob of N-in:
0.01894
outside
1 - 278
Population Genetic Test Statistics
Pi
225.273353
Theta
142.956258
Tajima's D
1.826202
CLR
0
CSRT
0.857607119644018
Interpretation
Uncertain
