Gene
KWMTBOMO11333 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001976
Annotation
Hemocyte_protein-glutamine_gamma-glutamyltransferase_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.868
Sequence
CDS
ATGGAGTCTCTACTTGTTGACGTTGTACACTTCTACCCACGAGAAAATGCTCAACCTCATAATACTATCAGATACGAGGTCGTAAACGATGAGGTCCCGACAACTACGATCGTCCGTCGCGGCCAGCCTTTTAACGGGGTCATCCGGTTCACCAGACCCTACGACGAGACGAATGACCTAGTGCAACTAGTCTTCTCGCTTGGTGACAACCCCACAATGGAGACTCAAGGGTCGGTATTCCCGCGCCGTGATGCGGTAGCCGACAAACATTCATGGACCGCTAAACTAATAGACGTACAAGAAAACACACTATCCTTTGAGGTATCAACTCCTGTGACGCTTCCTGTGGGCACTTGGTCTCTCCGAGTAGTCACCAGACAGAAGAACTCATCAGCCCGCGAGATCTACGACTTTGACCAGGATATTTACATTCTTTTTAACCCATGGAATCCTGATGACCAAACCTACATGGAAGACTGCACTCTACTCCAGGAGTATGTGCTAAACGACGTTGGTAAGGTCTGGGTGGGTCCCATCAAGACGACCAGGGGAAAGCCGTGGTTCTATGGACAGTTTGACGCTGTCGTCCTACCCGCCTGTATGTTTATGCTGGATAAAGCCGAATTTCCTTTCAACCAACGCGGTGATCCAGTTAAGGTAGCTCGTGTTATTTCAAAGATTGTGAACTCGAATGACGATGACGGAGTATTGGCTGGTCGTTGGGATGGGCAGTACGACGATGGAACAGCCCCGTCCGAATGGACCGGTTCTGTGGAGATCCTGGCACAATTCCTGGAGACCCAGCAGACGGTGCCCTATGGGCAGTGCTGGGTGTTCGCTGGCGTCGTCACCGCCGTATGCAGAGCCTTGGGGATCCCATCACGAGTAGTGTCCAATTTAGTGTCGGCCCACGACGCTAACAGCACTCTGACGGTCGACAAATACTACAGCGAGAACATGGAAGAGATGGAATCCGTTGACTCCATCTGGAACTATCACGTGTGGAACGATGTTTGGATGGCGCGACCAGACCTCCCAACCGGTAAGTCTATAGCTGATGGGATAATGAAGGGGGGATGCAGTGAATTATTTGGAAAATAA
Protein
MESLLVDVVHFYPRENAQPHNTIRYEVVNDEVPTTTIVRRGQPFNGVIRFTRPYDETNDLVQLVFSLGDNPTMETQGSVFPRRDAVADKHSWTAKLIDVQENTLSFEVSTPVTLPVGTWSLRVVTRQKNSSAREIYDFDQDIYILFNPWNPDDQTYMEDCTLLQEYVLNDVGKVWVGPIKTTRGKPWFYGQFDAVVLPACMFMLDKAEFPFNQRGDPVKVARVISKIVNSNDDDGVLAGRWDGQYDDGTAPSEWTGSVEILAQFLETQQTVPYGQCWVFAGVVTAVCRALGIPSRVVSNLVSAHDANSTLTVDKYYSENMEEMESVDSIWNYHVWNDVWMARPDLPTGKSIADGIMKGGCSELFGK
Summary
Cofactor
Ca(2+)
Uniprot
A0A194QFA5
A0A0N0PA38
A0A194R102
H9IXJ3
A0A2H1W2Z2
A0A212FNQ8
+ More
A0A1W4WI76 A0A0J7KRA2 A0A3L8DGG8 E2AYU1 A0A0L7RFM3 A0A088AKR6 A0A2A3ERM9 E9ID81 A0A0M9A5H6 A0A154P0F6 A0A195FLR1 A0A195BZU0 A0A158NEJ8 A0A151XIU9 A0A195BUU7 D6WH00 A0A026WXV5 A0A310SIN2 A0A1B6KC33 E2B2K7 F4WRW8 A0A1B6J0F0 A0A1B6D4M4 A0A1B6EUM3 A0A2J7PV97 A0A0C9QRW2 A0A1B6FF90 A0A0P5KUW0 A0A0P5NLU1 A0A0N8AW90 A0A0P6EC35 A0A0P6F7B3 A0A0P5GU29 A0A0P5GYP7 A0A0P6FKQ0 A0A0N8BSX9 A0A0P5PBD2 A0A0P6GNN4 A0A0P5I9M1 A0A0P6FAB0 A0A0P5R3L8 A0A0P5QG12 A0A0P6H070 A0A0P5RR98 A0A0P5CG03 A0A0P5S554 A0A0P6HJB8 A0A0P5KKD8 A0A0P5S7V1 A0A0P5I5T4 A0A0P5L885 A0A0P5RR37 A0A0P4XIZ5 A0A0P5JWC9 A0A0P5JMT5 A0A0P6CYF2 A0A0P5CIL7 A0A0P5ZXX2 A0A0P4YUC5 A0A0P4Z368 E9FR81 A0A067RI89 A0A0P5E099 A0A0P5YQW1 A0A0N7ZQR4 A0A2P8YH94 A0A0P6FYM4 A0A0P5ML04 T1HFP9 A0A162DAJ7 A0A0P6IF44 A0A067RCZ6 A0A0P5XBZ4 A0A0P5VMP7 A0A1D2N8Q4 A0A0N8AAG4 A0A0P6DKR4 A0A0P6C2U9 A0A1B6CDA4 A0A224XJ93 A0A226F4B1 A0A0P5WBH8 J9K0Q3 A0A2S2NU79 A0A023EZ45 A0A2P2HW15 A0A2J7QXL3 A0A0V0G4Q1 Q68Y89 A0A147BWD1 A0A131Y2F0
A0A1W4WI76 A0A0J7KRA2 A0A3L8DGG8 E2AYU1 A0A0L7RFM3 A0A088AKR6 A0A2A3ERM9 E9ID81 A0A0M9A5H6 A0A154P0F6 A0A195FLR1 A0A195BZU0 A0A158NEJ8 A0A151XIU9 A0A195BUU7 D6WH00 A0A026WXV5 A0A310SIN2 A0A1B6KC33 E2B2K7 F4WRW8 A0A1B6J0F0 A0A1B6D4M4 A0A1B6EUM3 A0A2J7PV97 A0A0C9QRW2 A0A1B6FF90 A0A0P5KUW0 A0A0P5NLU1 A0A0N8AW90 A0A0P6EC35 A0A0P6F7B3 A0A0P5GU29 A0A0P5GYP7 A0A0P6FKQ0 A0A0N8BSX9 A0A0P5PBD2 A0A0P6GNN4 A0A0P5I9M1 A0A0P6FAB0 A0A0P5R3L8 A0A0P5QG12 A0A0P6H070 A0A0P5RR98 A0A0P5CG03 A0A0P5S554 A0A0P6HJB8 A0A0P5KKD8 A0A0P5S7V1 A0A0P5I5T4 A0A0P5L885 A0A0P5RR37 A0A0P4XIZ5 A0A0P5JWC9 A0A0P5JMT5 A0A0P6CYF2 A0A0P5CIL7 A0A0P5ZXX2 A0A0P4YUC5 A0A0P4Z368 E9FR81 A0A067RI89 A0A0P5E099 A0A0P5YQW1 A0A0N7ZQR4 A0A2P8YH94 A0A0P6FYM4 A0A0P5ML04 T1HFP9 A0A162DAJ7 A0A0P6IF44 A0A067RCZ6 A0A0P5XBZ4 A0A0P5VMP7 A0A1D2N8Q4 A0A0N8AAG4 A0A0P6DKR4 A0A0P6C2U9 A0A1B6CDA4 A0A224XJ93 A0A226F4B1 A0A0P5WBH8 J9K0Q3 A0A2S2NU79 A0A023EZ45 A0A2P2HW15 A0A2J7QXL3 A0A0V0G4Q1 Q68Y89 A0A147BWD1 A0A131Y2F0
Pubmed
EMBL
KQ459053
KPJ04094.1
KQ458864
KPJ04688.1
KQ461073
KPJ09521.1
+ More
BABH01018249 ODYU01006003 SOQ47465.1 AGBW02004770 OWR55378.1 LBMM01004039 KMQ92873.1 QOIP01000008 RLU19411.1 GL444002 EFN61386.1 KQ414599 KOC69777.1 KZ288195 PBC33779.1 GL762454 EFZ21401.1 KQ435750 KOX76269.1 KQ434791 KZC05406.1 KQ981490 KYN41187.1 KQ978501 KYM93428.1 ADTU01013469 KQ982080 KYQ60333.1 KQ976401 KYM92394.1 KQ971321 EFA00611.1 KK107064 EZA60872.1 KQ765323 OAD53974.1 GEBQ01031002 JAT08975.1 GL445147 EFN90084.1 GL888292 EGI63125.1 GECU01015080 JAS92626.1 GEDC01016678 JAS20620.1 GECZ01028113 JAS41656.1 NEVH01020980 PNF20253.1 GBYB01003366 GBYB01003367 JAG73133.1 JAG73134.1 GECZ01020887 JAS48882.1 GDIQ01180362 JAK71363.1 GDIQ01140518 JAL11208.1 GDIQ01236739 JAK14986.1 GDIQ01064856 JAN29881.1 GDIQ01051765 JAN42972.1 GDIQ01235593 JAK16132.1 GDIQ01234436 JAK17289.1 GDIQ01049001 JAN45736.1 GDIQ01148027 JAL03699.1 GDIQ01130981 JAL20745.1 GDIQ01050407 JAN44330.1 GDIQ01217943 JAK33782.1 GDIQ01066206 JAN28531.1 GDIQ01105938 JAL45788.1 GDIQ01114679 JAL37047.1 GDIQ01047454 JAN47283.1 GDIQ01097137 JAL54589.1 GDIP01171083 GDIP01145781 GDIP01125646 JAJ52319.1 GDIQ01098638 JAL53088.1 GDIQ01032163 JAN62574.1 GDIQ01182807 JAK68918.1 GDIQ01100040 JAL51686.1 GDIQ01219781 JAK31944.1 GDIQ01178749 JAK72976.1 GDIQ01098639 JAL53087.1 GDIP01240538 JAI82863.1 GDIQ01198497 JAK53228.1 GDIQ01200953 JAK50772.1 GDIQ01085751 JAN08986.1 GDIP01171084 JAJ52318.1 GDIP01041635 JAM62080.1 GDIP01222968 JAJ00434.1 GDIP01230771 JAI92630.1 GL732523 EFX90239.1 KK852496 KDR22723.1 GDIP01166624 JAJ56778.1 GDIP01054548 JAM49167.1 GDIP01221825 JAJ01577.1 PYGN01000598 PSN43603.1 GDIQ01051766 JAN42971.1 GDIQ01162160 JAK89565.1 ACPB03003823 LRGB01002121 KZS09124.1 GDIQ01005478 JAN89259.1 KK852781 KDR16648.1 GDIP01074345 JAM29370.1 GDIP01097945 JAM05770.1 LJIJ01000150 ODN01465.1 GDIP01166625 JAJ56777.1 GDIQ01090929 JAN03808.1 GDIP01021961 JAM81754.1 GEDC01025859 JAS11439.1 GFTR01007764 JAW08662.1 LNIX01000001 OXA63756.1 GDIP01089546 JAM14169.1 ABLF02022376 ABLF02022379 GGMR01008110 MBY20729.1 GBBI01004224 JAC14488.1 IACF01000182 LAB65985.1 NEVH01009379 PNF33318.1 GECL01003105 JAP03019.1 AB162767 BAD36808.1 GEGO01000255 JAR95149.1 GEFM01002377 JAP73419.1
BABH01018249 ODYU01006003 SOQ47465.1 AGBW02004770 OWR55378.1 LBMM01004039 KMQ92873.1 QOIP01000008 RLU19411.1 GL444002 EFN61386.1 KQ414599 KOC69777.1 KZ288195 PBC33779.1 GL762454 EFZ21401.1 KQ435750 KOX76269.1 KQ434791 KZC05406.1 KQ981490 KYN41187.1 KQ978501 KYM93428.1 ADTU01013469 KQ982080 KYQ60333.1 KQ976401 KYM92394.1 KQ971321 EFA00611.1 KK107064 EZA60872.1 KQ765323 OAD53974.1 GEBQ01031002 JAT08975.1 GL445147 EFN90084.1 GL888292 EGI63125.1 GECU01015080 JAS92626.1 GEDC01016678 JAS20620.1 GECZ01028113 JAS41656.1 NEVH01020980 PNF20253.1 GBYB01003366 GBYB01003367 JAG73133.1 JAG73134.1 GECZ01020887 JAS48882.1 GDIQ01180362 JAK71363.1 GDIQ01140518 JAL11208.1 GDIQ01236739 JAK14986.1 GDIQ01064856 JAN29881.1 GDIQ01051765 JAN42972.1 GDIQ01235593 JAK16132.1 GDIQ01234436 JAK17289.1 GDIQ01049001 JAN45736.1 GDIQ01148027 JAL03699.1 GDIQ01130981 JAL20745.1 GDIQ01050407 JAN44330.1 GDIQ01217943 JAK33782.1 GDIQ01066206 JAN28531.1 GDIQ01105938 JAL45788.1 GDIQ01114679 JAL37047.1 GDIQ01047454 JAN47283.1 GDIQ01097137 JAL54589.1 GDIP01171083 GDIP01145781 GDIP01125646 JAJ52319.1 GDIQ01098638 JAL53088.1 GDIQ01032163 JAN62574.1 GDIQ01182807 JAK68918.1 GDIQ01100040 JAL51686.1 GDIQ01219781 JAK31944.1 GDIQ01178749 JAK72976.1 GDIQ01098639 JAL53087.1 GDIP01240538 JAI82863.1 GDIQ01198497 JAK53228.1 GDIQ01200953 JAK50772.1 GDIQ01085751 JAN08986.1 GDIP01171084 JAJ52318.1 GDIP01041635 JAM62080.1 GDIP01222968 JAJ00434.1 GDIP01230771 JAI92630.1 GL732523 EFX90239.1 KK852496 KDR22723.1 GDIP01166624 JAJ56778.1 GDIP01054548 JAM49167.1 GDIP01221825 JAJ01577.1 PYGN01000598 PSN43603.1 GDIQ01051766 JAN42971.1 GDIQ01162160 JAK89565.1 ACPB03003823 LRGB01002121 KZS09124.1 GDIQ01005478 JAN89259.1 KK852781 KDR16648.1 GDIP01074345 JAM29370.1 GDIP01097945 JAM05770.1 LJIJ01000150 ODN01465.1 GDIP01166625 JAJ56777.1 GDIQ01090929 JAN03808.1 GDIP01021961 JAM81754.1 GEDC01025859 JAS11439.1 GFTR01007764 JAW08662.1 LNIX01000001 OXA63756.1 GDIP01089546 JAM14169.1 ABLF02022376 ABLF02022379 GGMR01008110 MBY20729.1 GBBI01004224 JAC14488.1 IACF01000182 LAB65985.1 NEVH01009379 PNF33318.1 GECL01003105 JAP03019.1 AB162767 BAD36808.1 GEGO01000255 JAR95149.1 GEFM01002377 JAP73419.1
Proteomes
UP000053268
UP000053240
UP000005204
UP000007151
UP000192223
UP000036403
+ More
UP000279307 UP000000311 UP000053825 UP000005203 UP000242457 UP000053105 UP000076502 UP000078541 UP000078542 UP000005205 UP000075809 UP000078540 UP000007266 UP000053097 UP000008237 UP000007755 UP000235965 UP000000305 UP000027135 UP000245037 UP000015103 UP000076858 UP000094527 UP000198287 UP000007819
UP000279307 UP000000311 UP000053825 UP000005203 UP000242457 UP000053105 UP000076502 UP000078541 UP000078542 UP000005205 UP000075809 UP000078540 UP000007266 UP000053097 UP000008237 UP000007755 UP000235965 UP000000305 UP000027135 UP000245037 UP000015103 UP000076858 UP000094527 UP000198287 UP000007819
PRIDE
Interpro
IPR008958
Transglutaminase_C
+ More
IPR036238 Transglutaminase_C_sf
IPR002931 Transglutaminase-like
IPR036985 Transglutaminase-like_sf
IPR001102 Transglutaminase_N
IPR013808 Transglutaminase_AS
IPR014756 Ig_E-set
IPR006612 THAP_Znf
IPR013783 Ig-like_fold
IPR038765 Papain-like_cys_pep_sf
IPR023608 Transglutaminase_animal
IPR036238 Transglutaminase_C_sf
IPR002931 Transglutaminase-like
IPR036985 Transglutaminase-like_sf
IPR001102 Transglutaminase_N
IPR013808 Transglutaminase_AS
IPR014756 Ig_E-set
IPR006612 THAP_Znf
IPR013783 Ig-like_fold
IPR038765 Papain-like_cys_pep_sf
IPR023608 Transglutaminase_animal
Gene 3D
ProteinModelPortal
A0A194QFA5
A0A0N0PA38
A0A194R102
H9IXJ3
A0A2H1W2Z2
A0A212FNQ8
+ More
A0A1W4WI76 A0A0J7KRA2 A0A3L8DGG8 E2AYU1 A0A0L7RFM3 A0A088AKR6 A0A2A3ERM9 E9ID81 A0A0M9A5H6 A0A154P0F6 A0A195FLR1 A0A195BZU0 A0A158NEJ8 A0A151XIU9 A0A195BUU7 D6WH00 A0A026WXV5 A0A310SIN2 A0A1B6KC33 E2B2K7 F4WRW8 A0A1B6J0F0 A0A1B6D4M4 A0A1B6EUM3 A0A2J7PV97 A0A0C9QRW2 A0A1B6FF90 A0A0P5KUW0 A0A0P5NLU1 A0A0N8AW90 A0A0P6EC35 A0A0P6F7B3 A0A0P5GU29 A0A0P5GYP7 A0A0P6FKQ0 A0A0N8BSX9 A0A0P5PBD2 A0A0P6GNN4 A0A0P5I9M1 A0A0P6FAB0 A0A0P5R3L8 A0A0P5QG12 A0A0P6H070 A0A0P5RR98 A0A0P5CG03 A0A0P5S554 A0A0P6HJB8 A0A0P5KKD8 A0A0P5S7V1 A0A0P5I5T4 A0A0P5L885 A0A0P5RR37 A0A0P4XIZ5 A0A0P5JWC9 A0A0P5JMT5 A0A0P6CYF2 A0A0P5CIL7 A0A0P5ZXX2 A0A0P4YUC5 A0A0P4Z368 E9FR81 A0A067RI89 A0A0P5E099 A0A0P5YQW1 A0A0N7ZQR4 A0A2P8YH94 A0A0P6FYM4 A0A0P5ML04 T1HFP9 A0A162DAJ7 A0A0P6IF44 A0A067RCZ6 A0A0P5XBZ4 A0A0P5VMP7 A0A1D2N8Q4 A0A0N8AAG4 A0A0P6DKR4 A0A0P6C2U9 A0A1B6CDA4 A0A224XJ93 A0A226F4B1 A0A0P5WBH8 J9K0Q3 A0A2S2NU79 A0A023EZ45 A0A2P2HW15 A0A2J7QXL3 A0A0V0G4Q1 Q68Y89 A0A147BWD1 A0A131Y2F0
A0A1W4WI76 A0A0J7KRA2 A0A3L8DGG8 E2AYU1 A0A0L7RFM3 A0A088AKR6 A0A2A3ERM9 E9ID81 A0A0M9A5H6 A0A154P0F6 A0A195FLR1 A0A195BZU0 A0A158NEJ8 A0A151XIU9 A0A195BUU7 D6WH00 A0A026WXV5 A0A310SIN2 A0A1B6KC33 E2B2K7 F4WRW8 A0A1B6J0F0 A0A1B6D4M4 A0A1B6EUM3 A0A2J7PV97 A0A0C9QRW2 A0A1B6FF90 A0A0P5KUW0 A0A0P5NLU1 A0A0N8AW90 A0A0P6EC35 A0A0P6F7B3 A0A0P5GU29 A0A0P5GYP7 A0A0P6FKQ0 A0A0N8BSX9 A0A0P5PBD2 A0A0P6GNN4 A0A0P5I9M1 A0A0P6FAB0 A0A0P5R3L8 A0A0P5QG12 A0A0P6H070 A0A0P5RR98 A0A0P5CG03 A0A0P5S554 A0A0P6HJB8 A0A0P5KKD8 A0A0P5S7V1 A0A0P5I5T4 A0A0P5L885 A0A0P5RR37 A0A0P4XIZ5 A0A0P5JWC9 A0A0P5JMT5 A0A0P6CYF2 A0A0P5CIL7 A0A0P5ZXX2 A0A0P4YUC5 A0A0P4Z368 E9FR81 A0A067RI89 A0A0P5E099 A0A0P5YQW1 A0A0N7ZQR4 A0A2P8YH94 A0A0P6FYM4 A0A0P5ML04 T1HFP9 A0A162DAJ7 A0A0P6IF44 A0A067RCZ6 A0A0P5XBZ4 A0A0P5VMP7 A0A1D2N8Q4 A0A0N8AAG4 A0A0P6DKR4 A0A0P6C2U9 A0A1B6CDA4 A0A224XJ93 A0A226F4B1 A0A0P5WBH8 J9K0Q3 A0A2S2NU79 A0A023EZ45 A0A2P2HW15 A0A2J7QXL3 A0A0V0G4Q1 Q68Y89 A0A147BWD1 A0A131Y2F0
PDB
5MHO
E-value=6.69538e-71,
Score=679
Ontologies
GO
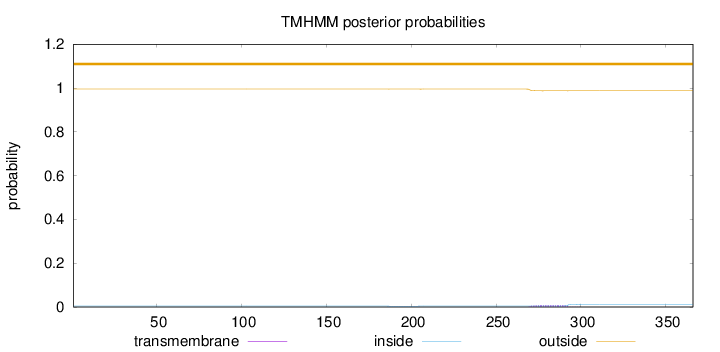
Topology
Length:
366
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.21217
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00405
outside
1 - 366
Population Genetic Test Statistics
Pi
228.301298
Theta
167.924854
Tajima's D
1.47296
CLR
0.40263
CSRT
0.776661166941653
Interpretation
Uncertain
