Gene
KWMTBOMO11332 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001976
Annotation
PREDICTED:_hemocyte_protein-glutamine_gamma-glutamyltransferase-like_[Amyelois_transitella]
Full name
Hemocyte protein-glutamine gamma-glutamyltransferase
Alternative Name
Hemocyte transglutaminase
Location in the cell
Cytoplasmic Reliability : 1.602
Sequence
CDS
ATGGTGCTGACGAAGAAGCCGTTCGTGTTTGACCCGATCGGCGATGAAGACCGGGAAGACATCACCAGCCAGTACAAATACAAAGAAGGCACGGCTTCAGAGAGGCTGGCCCTGATGAACGGAGTCAGATATTCGGAGAGAGCGAAGAGGTACTACTCAGTAGTGAGCAACTTACAGACGGACGTGACCTTCAAACTAAGAGACATCGATTTAGTTTCCATCGGCAAAGACTTCAAGGTCACAATCGACATCAAGAATGTTTCGGATCAGGGACGTAACATAAAGGCATCATTGAGCGCATCGAGTTTCTACTACAACGGAGTTAAATTTGACATCATCAAAAAAATCGAAGGAAAACTCTACGTTGGACCACAGAAAAACGAAGAAGTCAGCCTCGTGGTGAAAGTAGAGGACTATTTGCCGAAGCTGGTGGAGTACGGGAACATCAAGATATCTGCGATGGCCATTGTCGATGAAACGAAGCAGTCCTGGGCTGACGACGACGACTTCCAGATCCTCAAACCGAACATTAACATTAAGTTCAAAGACGACCTGATCATCGGGCAGCCGGCAACAGTGGAACTGTCGCTTTTGAACCCTCTGAACAGTACGCTGAGTGGGTGCGAGTTCCGCGTGACGTCATCGGGAATAGCGGGTCGTGCCCTGCGCGTTCCCGCCGCGGACGTCGCTCCTGAAGGACTACTCACCGTTGAACTACCTATTCAACCTAACAAACTGGGTAATATCAGTTTCGTGGCCACATTCAAGTCGAACGAGCTGAAGGACATCAACGGAGCCGCCACCACGGAAGTGCTCGAAGGGTAA
Protein
MVLTKKPFVFDPIGDEDREDITSQYKYKEGTASERLALMNGVRYSERAKRYYSVVSNLQTDVTFKLRDIDLVSIGKDFKVTIDIKNVSDQGRNIKASLSASSFYYNGVKFDIIKKIEGKLYVGPQKNEEVSLVVKVEDYLPKLVEYGNIKISAMAIVDETKQSWADDDDFQILKPNINIKFKDDLIIGQPATVELSLLNPLNSTLSGCEFRVTSSGIAGRALRVPAADVAPEGLLTVELPIQPNKLGNISFVATFKSNELKDINGAATTEVLEG
Summary
Catalytic Activity
L-glutaminyl-[protein] + L-lysyl-[protein] = [protein]-L-lysyl-N(6)-5-L-glutamyl-[protein] + NH4(+)
Cofactor
Ca(2+)
Similarity
Belongs to the transglutaminase superfamily. Transglutaminase family.
Keywords
Acyltransferase
Calcium
Direct protein sequencing
Membrane
Metal-binding
Transferase
Feature
chain Hemocyte protein-glutamine gamma-glutamyltransferase
Uniprot
H9IXJ3
A0A3S2LTZ0
A0A2W1BNN0
A0A212FNQ8
A0A194R102
A0A194QFA5
+ More
A0A2H1W2Z2 A0A0L7KZY8 A0A0J7KRL0 A0A2J7PV97 F4WRW8 A0A195FLR1 A0A151XIU9 A0A195BZU0 A0A158NEJ8 E2AYU1 E9ID81 A0A195BUU7 A0A154P0F6 E2B2K7 A0A0L7RFM3 A0A026WXV5 A0A3L8DGG8 A0A0M9A5H6 A0A310SIN2 A0A2A3ERM9 A0A088AKR6 A0A067RI89 A0A0C9QRW2 A0A1Y1KRA9 D6WH00 A0A1B6EUM3 A0A1Y1KS20 A0A1B6D4M4 A0A1W4WI76 A0A1B6KC33 A0A195EN57 A0A2P8YH94 A0A0P5LX05 A0A0P4XT55 A0A0N8C9J7 A0A0P5V592 A0A0P5B761 E9FR81 A0A0P5VIZ2 A0A162DAJ7 A0A0P6DKR4 A0A0P5WA68 A0A0P5XBZ4 A0A0N8AAG4 A0A0P6C2U9 A0A1B6I894 A0A0P5VMP7 A0A1B6K5T3 A0A023EZ45 A0A0P5ML04 Q05187 A0A224XJ93 A0A0V0G4Q1 A0A023F0E9 A0A0V0G7A9 A0A1B6H930 A0A2P2HW15 T1HFS7 A0A224XI60 A0A1D2N8Q4 A0A023EZ44 A0A0V0G6K0 T1J480 A0A0P6IF44 A0A2P6L0I5 A0A023FM45 T1I8S6 A0A023F227 A0A3R7SJE1 A0A2J7QXL3 T1HFP9 A0A224XFQ6 A0A0K8TIJ3 A0A146LM26 A0A0M3LC01 A0A1E1X9T3 A0A0A9WHI8 A0A444SNA9 A0A131Y2F0 A0A147BWD1 A0A444ST60 B7PSF8 A0A131XKN6 A0A0K2TPI8 A0A0K2TRC0 A0A131YNM1 T1HFV3 A3DT47 A0A023GNS4 A0A087T1V1 A0A212ESV0 A0A224YZS7
A0A2H1W2Z2 A0A0L7KZY8 A0A0J7KRL0 A0A2J7PV97 F4WRW8 A0A195FLR1 A0A151XIU9 A0A195BZU0 A0A158NEJ8 E2AYU1 E9ID81 A0A195BUU7 A0A154P0F6 E2B2K7 A0A0L7RFM3 A0A026WXV5 A0A3L8DGG8 A0A0M9A5H6 A0A310SIN2 A0A2A3ERM9 A0A088AKR6 A0A067RI89 A0A0C9QRW2 A0A1Y1KRA9 D6WH00 A0A1B6EUM3 A0A1Y1KS20 A0A1B6D4M4 A0A1W4WI76 A0A1B6KC33 A0A195EN57 A0A2P8YH94 A0A0P5LX05 A0A0P4XT55 A0A0N8C9J7 A0A0P5V592 A0A0P5B761 E9FR81 A0A0P5VIZ2 A0A162DAJ7 A0A0P6DKR4 A0A0P5WA68 A0A0P5XBZ4 A0A0N8AAG4 A0A0P6C2U9 A0A1B6I894 A0A0P5VMP7 A0A1B6K5T3 A0A023EZ45 A0A0P5ML04 Q05187 A0A224XJ93 A0A0V0G4Q1 A0A023F0E9 A0A0V0G7A9 A0A1B6H930 A0A2P2HW15 T1HFS7 A0A224XI60 A0A1D2N8Q4 A0A023EZ44 A0A0V0G6K0 T1J480 A0A0P6IF44 A0A2P6L0I5 A0A023FM45 T1I8S6 A0A023F227 A0A3R7SJE1 A0A2J7QXL3 T1HFP9 A0A224XFQ6 A0A0K8TIJ3 A0A146LM26 A0A0M3LC01 A0A1E1X9T3 A0A0A9WHI8 A0A444SNA9 A0A131Y2F0 A0A147BWD1 A0A444ST60 B7PSF8 A0A131XKN6 A0A0K2TPI8 A0A0K2TRC0 A0A131YNM1 T1HFV3 A3DT47 A0A023GNS4 A0A087T1V1 A0A212ESV0 A0A224YZS7
EC Number
2.3.2.13
Pubmed
EMBL
BABH01018249
RSAL01000205
RVE44370.1
KZ150054
PZC74340.1
AGBW02004770
+ More
OWR55378.1 KQ461073 KPJ09521.1 KQ459053 KPJ04094.1 ODYU01006003 SOQ47465.1 JTDY01004110 KOB68589.1 LBMM01004039 KMQ92874.1 NEVH01020980 PNF20253.1 GL888292 EGI63125.1 KQ981490 KYN41187.1 KQ982080 KYQ60333.1 KQ978501 KYM93428.1 ADTU01013469 GL444002 EFN61386.1 GL762454 EFZ21401.1 KQ976401 KYM92394.1 KQ434791 KZC05406.1 GL445147 EFN90084.1 KQ414599 KOC69777.1 KK107064 EZA60872.1 QOIP01000008 RLU19411.1 KQ435750 KOX76269.1 KQ765323 OAD53974.1 KZ288195 PBC33779.1 KK852496 KDR22723.1 GBYB01003366 GBYB01003367 JAG73133.1 JAG73134.1 GEZM01080273 JAV62235.1 KQ971321 EFA00611.1 GECZ01028113 JAS41656.1 GEZM01080272 JAV62236.1 GEDC01016678 JAS20620.1 GEBQ01031002 JAT08975.1 KQ978625 KYN29601.1 PYGN01000598 PSN43603.1 GDIQ01172949 JAK78776.1 GDIP01238123 JAI85278.1 GDIQ01101483 JAL50243.1 GDIP01104828 JAL98886.1 GDIP01188523 JAJ34879.1 GL732523 EFX90239.1 GDIP01099331 JAM04384.1 LRGB01002121 KZS09124.1 GDIQ01090929 JAN03808.1 GDIP01089547 JAM14168.1 GDIP01074345 JAM29370.1 GDIP01166625 JAJ56777.1 GDIP01021961 JAM81754.1 GECU01024577 JAS83129.1 GDIP01097945 JAM05770.1 GECU01000901 JAT06806.1 GBBI01004224 JAC14488.1 GDIQ01162160 JAK89565.1 D12593 GFTR01007764 JAW08662.1 GECL01003105 JAP03019.1 GBBI01004283 JAC14429.1 GECL01002238 JAP03886.1 GECU01036546 JAS71160.1 IACF01000182 LAB65985.1 ACPB03003823 GFTR01006928 JAW09498.1 LJIJ01000150 ODN01465.1 GBBI01004225 JAC14487.1 GECL01002393 JAP03731.1 JH431841 GDIQ01005478 JAN89259.1 MWRG01002934 PRD32083.1 GBBK01002203 JAC22279.1 ACPB03014010 GBBI01003591 JAC15121.1 QCYY01003436 ROT63475.1 NEVH01009379 PNF33318.1 GFTR01007788 JAW08638.1 GBRD01000413 JAG65408.1 GDHC01010364 JAQ08265.1 KM008611 AIN46597.1 GFAC01003337 JAT95851.1 GBHO01037631 JAG05973.1 SAUD01029459 RXG54888.1 GEFM01002377 JAP73419.1 GEGO01000255 JAR95149.1 SAUD01024053 RXG56573.1 ABJB010058208 ABJB010167361 ABJB010618350 ABJB010929525 DS778349 EEC09530.1 GEFH01002490 JAP66091.1 HACA01010567 CDW27928.1 HACA01010570 CDW27931.1 GEDV01008527 JAP80030.1 ACPB03003820 DQ318855 ABC33914.1 GBBM01000535 JAC34883.1 KK113013 KFM59090.1 AGBW02012723 OWR44549.1 GFPF01008338 MAA19484.1
OWR55378.1 KQ461073 KPJ09521.1 KQ459053 KPJ04094.1 ODYU01006003 SOQ47465.1 JTDY01004110 KOB68589.1 LBMM01004039 KMQ92874.1 NEVH01020980 PNF20253.1 GL888292 EGI63125.1 KQ981490 KYN41187.1 KQ982080 KYQ60333.1 KQ978501 KYM93428.1 ADTU01013469 GL444002 EFN61386.1 GL762454 EFZ21401.1 KQ976401 KYM92394.1 KQ434791 KZC05406.1 GL445147 EFN90084.1 KQ414599 KOC69777.1 KK107064 EZA60872.1 QOIP01000008 RLU19411.1 KQ435750 KOX76269.1 KQ765323 OAD53974.1 KZ288195 PBC33779.1 KK852496 KDR22723.1 GBYB01003366 GBYB01003367 JAG73133.1 JAG73134.1 GEZM01080273 JAV62235.1 KQ971321 EFA00611.1 GECZ01028113 JAS41656.1 GEZM01080272 JAV62236.1 GEDC01016678 JAS20620.1 GEBQ01031002 JAT08975.1 KQ978625 KYN29601.1 PYGN01000598 PSN43603.1 GDIQ01172949 JAK78776.1 GDIP01238123 JAI85278.1 GDIQ01101483 JAL50243.1 GDIP01104828 JAL98886.1 GDIP01188523 JAJ34879.1 GL732523 EFX90239.1 GDIP01099331 JAM04384.1 LRGB01002121 KZS09124.1 GDIQ01090929 JAN03808.1 GDIP01089547 JAM14168.1 GDIP01074345 JAM29370.1 GDIP01166625 JAJ56777.1 GDIP01021961 JAM81754.1 GECU01024577 JAS83129.1 GDIP01097945 JAM05770.1 GECU01000901 JAT06806.1 GBBI01004224 JAC14488.1 GDIQ01162160 JAK89565.1 D12593 GFTR01007764 JAW08662.1 GECL01003105 JAP03019.1 GBBI01004283 JAC14429.1 GECL01002238 JAP03886.1 GECU01036546 JAS71160.1 IACF01000182 LAB65985.1 ACPB03003823 GFTR01006928 JAW09498.1 LJIJ01000150 ODN01465.1 GBBI01004225 JAC14487.1 GECL01002393 JAP03731.1 JH431841 GDIQ01005478 JAN89259.1 MWRG01002934 PRD32083.1 GBBK01002203 JAC22279.1 ACPB03014010 GBBI01003591 JAC15121.1 QCYY01003436 ROT63475.1 NEVH01009379 PNF33318.1 GFTR01007788 JAW08638.1 GBRD01000413 JAG65408.1 GDHC01010364 JAQ08265.1 KM008611 AIN46597.1 GFAC01003337 JAT95851.1 GBHO01037631 JAG05973.1 SAUD01029459 RXG54888.1 GEFM01002377 JAP73419.1 GEGO01000255 JAR95149.1 SAUD01024053 RXG56573.1 ABJB010058208 ABJB010167361 ABJB010618350 ABJB010929525 DS778349 EEC09530.1 GEFH01002490 JAP66091.1 HACA01010567 CDW27928.1 HACA01010570 CDW27931.1 GEDV01008527 JAP80030.1 ACPB03003820 DQ318855 ABC33914.1 GBBM01000535 JAC34883.1 KK113013 KFM59090.1 AGBW02012723 OWR44549.1 GFPF01008338 MAA19484.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000036403 UP000235965 UP000007755 UP000078541 UP000075809 UP000078542 UP000005205 UP000000311 UP000078540 UP000076502 UP000008237 UP000053825 UP000053097 UP000279307 UP000053105 UP000242457 UP000005203 UP000027135 UP000007266 UP000192223 UP000078492 UP000245037 UP000000305 UP000076858 UP000015103 UP000094527 UP000283509 UP000288706 UP000001555 UP000054359
UP000036403 UP000235965 UP000007755 UP000078541 UP000075809 UP000078542 UP000005205 UP000000311 UP000078540 UP000076502 UP000008237 UP000053825 UP000053097 UP000279307 UP000053105 UP000242457 UP000005203 UP000027135 UP000007266 UP000192223 UP000078492 UP000245037 UP000000305 UP000076858 UP000015103 UP000094527 UP000283509 UP000288706 UP000001555 UP000054359
Interpro
IPR014756
Ig_E-set
+ More
IPR023608 Transglutaminase_animal
IPR036985 Transglutaminase-like_sf
IPR002931 Transglutaminase-like
IPR038765 Papain-like_cys_pep_sf
IPR013783 Ig-like_fold
IPR001102 Transglutaminase_N
IPR036238 Transglutaminase_C_sf
IPR008958 Transglutaminase_C
IPR013808 Transglutaminase_AS
IPR006612 THAP_Znf
IPR023608 Transglutaminase_animal
IPR036985 Transglutaminase-like_sf
IPR002931 Transglutaminase-like
IPR038765 Papain-like_cys_pep_sf
IPR013783 Ig-like_fold
IPR001102 Transglutaminase_N
IPR036238 Transglutaminase_C_sf
IPR008958 Transglutaminase_C
IPR013808 Transglutaminase_AS
IPR006612 THAP_Znf
Gene 3D
ProteinModelPortal
H9IXJ3
A0A3S2LTZ0
A0A2W1BNN0
A0A212FNQ8
A0A194R102
A0A194QFA5
+ More
A0A2H1W2Z2 A0A0L7KZY8 A0A0J7KRL0 A0A2J7PV97 F4WRW8 A0A195FLR1 A0A151XIU9 A0A195BZU0 A0A158NEJ8 E2AYU1 E9ID81 A0A195BUU7 A0A154P0F6 E2B2K7 A0A0L7RFM3 A0A026WXV5 A0A3L8DGG8 A0A0M9A5H6 A0A310SIN2 A0A2A3ERM9 A0A088AKR6 A0A067RI89 A0A0C9QRW2 A0A1Y1KRA9 D6WH00 A0A1B6EUM3 A0A1Y1KS20 A0A1B6D4M4 A0A1W4WI76 A0A1B6KC33 A0A195EN57 A0A2P8YH94 A0A0P5LX05 A0A0P4XT55 A0A0N8C9J7 A0A0P5V592 A0A0P5B761 E9FR81 A0A0P5VIZ2 A0A162DAJ7 A0A0P6DKR4 A0A0P5WA68 A0A0P5XBZ4 A0A0N8AAG4 A0A0P6C2U9 A0A1B6I894 A0A0P5VMP7 A0A1B6K5T3 A0A023EZ45 A0A0P5ML04 Q05187 A0A224XJ93 A0A0V0G4Q1 A0A023F0E9 A0A0V0G7A9 A0A1B6H930 A0A2P2HW15 T1HFS7 A0A224XI60 A0A1D2N8Q4 A0A023EZ44 A0A0V0G6K0 T1J480 A0A0P6IF44 A0A2P6L0I5 A0A023FM45 T1I8S6 A0A023F227 A0A3R7SJE1 A0A2J7QXL3 T1HFP9 A0A224XFQ6 A0A0K8TIJ3 A0A146LM26 A0A0M3LC01 A0A1E1X9T3 A0A0A9WHI8 A0A444SNA9 A0A131Y2F0 A0A147BWD1 A0A444ST60 B7PSF8 A0A131XKN6 A0A0K2TPI8 A0A0K2TRC0 A0A131YNM1 T1HFV3 A3DT47 A0A023GNS4 A0A087T1V1 A0A212ESV0 A0A224YZS7
A0A2H1W2Z2 A0A0L7KZY8 A0A0J7KRL0 A0A2J7PV97 F4WRW8 A0A195FLR1 A0A151XIU9 A0A195BZU0 A0A158NEJ8 E2AYU1 E9ID81 A0A195BUU7 A0A154P0F6 E2B2K7 A0A0L7RFM3 A0A026WXV5 A0A3L8DGG8 A0A0M9A5H6 A0A310SIN2 A0A2A3ERM9 A0A088AKR6 A0A067RI89 A0A0C9QRW2 A0A1Y1KRA9 D6WH00 A0A1B6EUM3 A0A1Y1KS20 A0A1B6D4M4 A0A1W4WI76 A0A1B6KC33 A0A195EN57 A0A2P8YH94 A0A0P5LX05 A0A0P4XT55 A0A0N8C9J7 A0A0P5V592 A0A0P5B761 E9FR81 A0A0P5VIZ2 A0A162DAJ7 A0A0P6DKR4 A0A0P5WA68 A0A0P5XBZ4 A0A0N8AAG4 A0A0P6C2U9 A0A1B6I894 A0A0P5VMP7 A0A1B6K5T3 A0A023EZ45 A0A0P5ML04 Q05187 A0A224XJ93 A0A0V0G4Q1 A0A023F0E9 A0A0V0G7A9 A0A1B6H930 A0A2P2HW15 T1HFS7 A0A224XI60 A0A1D2N8Q4 A0A023EZ44 A0A0V0G6K0 T1J480 A0A0P6IF44 A0A2P6L0I5 A0A023FM45 T1I8S6 A0A023F227 A0A3R7SJE1 A0A2J7QXL3 T1HFP9 A0A224XFQ6 A0A0K8TIJ3 A0A146LM26 A0A0M3LC01 A0A1E1X9T3 A0A0A9WHI8 A0A444SNA9 A0A131Y2F0 A0A147BWD1 A0A444ST60 B7PSF8 A0A131XKN6 A0A0K2TPI8 A0A0K2TRC0 A0A131YNM1 T1HFV3 A3DT47 A0A023GNS4 A0A087T1V1 A0A212ESV0 A0A224YZS7
PDB
1EX0
E-value=6.82325e-15,
Score=194
Ontologies
GO
Topology
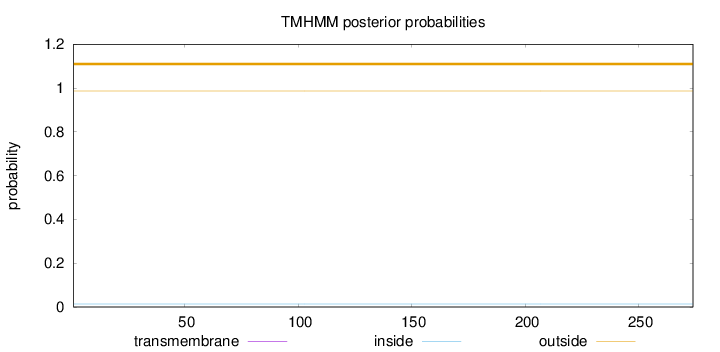
Subcellular location
Membrane
Length:
274
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00038
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01332
outside
1 - 274
Population Genetic Test Statistics
Pi
206.766838
Theta
183.390207
Tajima's D
0.763116
CLR
0.057071
CSRT
0.595020248987551
Interpretation
Uncertain
