Pre Gene Modal
BGIBMGA001847
Annotation
PREDICTED:_nicalin-1_[Papilio_polytes]
Full name
Nicalin
Location in the cell
PlasmaMembrane Reliability : 2.336
Sequence
CDS
ATGTGGCTGGATGAAGCAGACGGCTTTGCCGAAATATTCAAAGGCTATTTGCCTTTTTATCTTTTCGTTGCCCTACCTATTTTCATTATAATGTCTCCAGTGAATCCTGTGGCAGCCTCACACGAATTTTCCGTGTACAGAATGCAACAATATGACTTACACACAGTTCCCCATGGTTGTCGTAGTGCCAGCTTCAACTTAGAAGGTCGTTCTTTAACATCATGGAGCACATCACGTCACTGTGTCGTTGCACGCGTACAGGACATAACTTTAGAACAGTTCCTAGAAATCCGCAATAAGGCTGGGGCGTTGTTATTAGTATTACCTAAAAATGAAACTCTTTTGACGCCTGAAGAAAGAGAGCATATTCAATTGCTGGAAATGGCGATGGTACAACAAGAGATCAATGCACCCGTTTACTTTGCTCGTTGGACTCCTGAAATGGAGGACATTCTGGCTGACCTCCAGCATAGTTTTATTACGGATGACAAGTCAGGAACCGCTCTAGAGGCAATGTTCAACACTGTATCATCAAATGGATATCAAATCGTTGTCTCAGCCCCGACTCCACAGAAGCTCGATTCTAAACCAGTCACGTTACACGGCAAACTTATTGGGCGCTCTGGTTCAGCTCAAACCATTGTTATTGCTGCAAATTACGATTCCGCTGCCCTCGTACCGGAAATATCCCAAGGTGCAGATTGTAATGCATCAGGAGCGGTAGCATTACTGGAATTAGCCAGGATATTCTCGCGTATATACTCAACGGCTGGGGGGCGCGGAGCTCCGACTTTGGTCTTTGTGCTGACTTCGGTGGGGCACTCGCTCAACTATTTCGCCACTAAGAAATGGCTAGAAGAACAATTGGACTCCAGTGACGCTTCATTGCTTCAAGACGTGTCGTTCGTGAGCTGCATGGAGTGCGTGTCGCGCGGCCCGGTGCGCATGCACGTGTCCCGCCCCCCGCGCGCGCCCTCCCCCGCGCACTCCTTCCGCGCGCGGCTCGGGGCGCCGCTGCAGCACAAGAAGATCAACCTCGCCGACGAGCTGCTCGCCTGGCACCACGAGAGGTTCAGCATCAGGAGGATGACGGCCTTCACTCTTAGTTCATTGCAGAGTCACAAAGATTCAGGAAGGAGTACAGTGCTGGATACACCGTCTGAAGACAGAATCCAAAATTTAGTATCAAACGTGGCCAGGATAGCACGTGCTCTGGCCTCACATATTTACAACATCACTGATGATGAGAACGATGAGGATGCAGGTTTATATGATGATGTACTGAATGTAGATGAGGCAGCAATAAGATATTGGTACAATTATTTATCATCTCAATCAAGATCGGCTCACATAGTCACAACATCACCAAACGGAGGTGTGACCGGGGCATTAGAAAGAGTATTATCTAGATACATGGAGGTTACCGTGAGCACACACGCCGTCGATAAACGAGAGCCTGAGTATACTCTGTACTCACCCACTAGCGCCACCTTGTACGTTTACAGCGTTAAGCCGGCGGTCTTCGATCTCATTCTAACATTAGCGATAGTTTGTTATTTAGCTGTAGTTTATTTCGCGATACAAGCCTTCCCGCGTTTCTACGAAGAATACGCGAAGATCGTTACGGGCAAGACCAAAGTACAATAG
Protein
MWLDEADGFAEIFKGYLPFYLFVALPIFIIMSPVNPVAASHEFSVYRMQQYDLHTVPHGCRSASFNLEGRSLTSWSTSRHCVVARVQDITLEQFLEIRNKAGALLLVLPKNETLLTPEEREHIQLLEMAMVQQEINAPVYFARWTPEMEDILADLQHSFITDDKSGTALEAMFNTVSSNGYQIVVSAPTPQKLDSKPVTLHGKLIGRSGSAQTIVIAANYDSAALVPEISQGADCNASGAVALLELARIFSRIYSTAGGRGAPTLVFVLTSVGHSLNYFATKKWLEEQLDSSDASLLQDVSFVSCMECVSRGPVRMHVSRPPRAPSPAHSFRARLGAPLQHKKINLADELLAWHHERFSIRRMTAFTLSSLQSHKDSGRSTVLDTPSEDRIQNLVSNVARIARALASHIYNITDDENDEDAGLYDDVLNVDEAAIRYWYNYLSSQSRSAHIVTTSPNGGVTGALERVLSRYMEVTVSTHAVDKREPEYTLYSPTSATLYVYSVKPAVFDLILTLAIVCYLAVVYFAIQAFPRFYEEYAKIVTGKTKVQ
Summary
Description
May antagonize Nodal signaling and subsequent organization of axial structures during mesodermal patterning.
Similarity
Belongs to the nicastrin family.
Uniprot
A0A2H1W3D9
A0A194QEX6
A0A194QVM3
A0A2W1BJ60
H9IX64
A0A212FNS3
+ More
A0A437B1X3 S4NMA1 A0A1B6EWH8 A0A1B6LNK9 A0A026WTX8 A0A0L7KU27 A0A1Y1MDA6 K7IV63 A0A151IL08 E2AIS2 D6WSR0 A0A151WEL7 A0A151IVJ8 A0A195BES7 A0A158NNP9 A0A2J7QK95 A0A0C9RN96 A0A0L7RC92 A0A067R2G1 A0A310SF45 A0A1W4XB13 A0A087ZYD4 A0A154P742 F4X363 A0A195FH69 J3JUX1 A0A2A3E6I6 A0A0J7P331 A0A0A9XWJ6 A0A023F0P4 K1R1W9 A0A224XHN6 A0A2T7NEJ3 A0A2S2QJB6 A0A433TU00 A0A0L8H611 J9JX24 A0A023ETX0 A0A2S2PB97 T1JL86 A0A087UN08 A0A0P6F1L9 Q177H2 A0A147BV28 A0A131XZW9 C3YBA6 A0A0N8BB37 A0A0P4Y5M3 A0A0P5H2V8 A0A182FNY0 A0A2M4BJ54 A0A0P5VZE4 A0A182YHR2 A0A182XAW1 A0A182V7H0 W8BQR4 A0A182HSR1 Q7QCB7 A0A1Q3FAZ1 A0A1Q3FBK8 A0A131XCD4 A0A182U0M6 B0W936 A0A084VCX8 U5EYI1 E9FRQ1 A0A2M3Z7E3 A0A131YK52 A0A023FZZ2 A0A401SEA9 W5JIC9 V9KIE2 L7M6Z4 A0A182LET4 A0A1E1X949 A0A1E1XUB3 A0A023FPQ0 L7M878 A0A3B3ZSQ8 W5M914 V4CLX7 A0A1L8E295 A0A2M4AFL0 A0A2M4AFM9 A0A182M8Z5 A0A182NRM6 A0A401NQ32 A0A182R055 A0A182JYH5 A0A182RS58 A0A182VWV8
A0A437B1X3 S4NMA1 A0A1B6EWH8 A0A1B6LNK9 A0A026WTX8 A0A0L7KU27 A0A1Y1MDA6 K7IV63 A0A151IL08 E2AIS2 D6WSR0 A0A151WEL7 A0A151IVJ8 A0A195BES7 A0A158NNP9 A0A2J7QK95 A0A0C9RN96 A0A0L7RC92 A0A067R2G1 A0A310SF45 A0A1W4XB13 A0A087ZYD4 A0A154P742 F4X363 A0A195FH69 J3JUX1 A0A2A3E6I6 A0A0J7P331 A0A0A9XWJ6 A0A023F0P4 K1R1W9 A0A224XHN6 A0A2T7NEJ3 A0A2S2QJB6 A0A433TU00 A0A0L8H611 J9JX24 A0A023ETX0 A0A2S2PB97 T1JL86 A0A087UN08 A0A0P6F1L9 Q177H2 A0A147BV28 A0A131XZW9 C3YBA6 A0A0N8BB37 A0A0P4Y5M3 A0A0P5H2V8 A0A182FNY0 A0A2M4BJ54 A0A0P5VZE4 A0A182YHR2 A0A182XAW1 A0A182V7H0 W8BQR4 A0A182HSR1 Q7QCB7 A0A1Q3FAZ1 A0A1Q3FBK8 A0A131XCD4 A0A182U0M6 B0W936 A0A084VCX8 U5EYI1 E9FRQ1 A0A2M3Z7E3 A0A131YK52 A0A023FZZ2 A0A401SEA9 W5JIC9 V9KIE2 L7M6Z4 A0A182LET4 A0A1E1X949 A0A1E1XUB3 A0A023FPQ0 L7M878 A0A3B3ZSQ8 W5M914 V4CLX7 A0A1L8E295 A0A2M4AFL0 A0A2M4AFM9 A0A182M8Z5 A0A182NRM6 A0A401NQ32 A0A182R055 A0A182JYH5 A0A182RS58 A0A182VWV8
Pubmed
26354079
28756777
19121390
22118469
23622113
24508170
+ More
30249741 26227816 28004739 20075255 20798317 18362917 19820115 21347285 24845553 21719571 22516182 25401762 26823975 25474469 22992520 24945155 17510324 29652888 18563158 25244985 24495485 12364791 14747013 17210077 28049606 24438588 21292972 26830274 30297745 20920257 23761445 24402279 25576852 20966253 28503490 29209593 25463417 23254933
30249741 26227816 28004739 20075255 20798317 18362917 19820115 21347285 24845553 21719571 22516182 25401762 26823975 25474469 22992520 24945155 17510324 29652888 18563158 25244985 24495485 12364791 14747013 17210077 28049606 24438588 21292972 26830274 30297745 20920257 23761445 24402279 25576852 20966253 28503490 29209593 25463417 23254933
EMBL
ODYU01006003
SOQ47466.1
KQ459053
KPJ04093.1
KQ461073
KPJ09522.1
+ More
KZ150054 PZC74341.1 BABH01018251 BABH01018252 BABH01018253 BABH01018254 AGBW02004770 OWR55377.1 RSAL01000205 RVE44369.1 GAIX01014371 JAA78189.1 GECZ01027491 JAS42278.1 GEBQ01014771 JAT25206.1 KK107106 QOIP01000002 EZA59423.1 RLU25848.1 JTDY01005670 KOB66768.1 GEZM01036586 JAV82580.1 AAZX01001200 KQ977151 KYN05292.1 GL439874 EFN66661.1 KQ971354 EFA05879.1 KQ983238 KYQ46247.1 KQ980895 KYN11743.1 KQ976509 KYM82687.1 ADTU01000325 NEVH01013275 PNF28998.1 GBYB01008556 GBYB01008560 JAG78323.1 JAG78327.1 KQ414616 KOC68448.1 KK853060 KDR11903.1 KQ769179 OAD52925.1 KQ434829 KZC07749.1 GL888609 EGI59044.1 KQ981606 KYN39577.1 BT127036 AEE61998.1 KZ288369 PBC26779.1 LBMM01000238 KMR03323.1 GBHO01020351 GBHO01020349 GBHO01020348 GBRD01015144 GBRD01015143 GBRD01015142 GDHC01017288 JAG23253.1 JAG23255.1 JAG23256.1 JAG50682.1 JAQ01341.1 GBBI01003677 JAC15035.1 JH818095 EKC37504.1 GFTR01007108 JAW09318.1 PZQS01000013 PVD19594.1 GGMS01008646 MBY77849.1 RQTK01000184 RUS85048.1 KQ419065 KOF84726.1 ABLF02014212 GAPW01000800 JAC12798.1 GGMR01013839 MBY26458.1 JH432114 KK120653 KFM78747.1 GDIQ01054315 JAN40422.1 CH477376 EAT42305.1 GEGO01001012 JAR94392.1 GEFM01004520 JAP71276.1 GG666497 EEN62263.1 GDIQ01195163 JAK56562.1 GDIP01233283 LRGB01002568 JAI90118.1 KZS07100.1 GDIQ01233391 JAK18334.1 GGFJ01003958 MBW53099.1 GDIP01093053 JAM10662.1 GAMC01002950 JAC03606.1 APCN01000326 AAAB01008859 EAA07525.4 GFDL01010382 JAV24663.1 GFDL01010162 JAV24883.1 GEFH01004831 JAP63750.1 DS231862 EDS39733.1 ATLV01010990 KE524630 KFB35822.1 GANO01000284 JAB59587.1 GL732523 EFX90447.1 GGFM01003686 MBW24437.1 GEDV01008893 JAP79664.1 GBBL01000987 JAC26333.1 BEZZ01000215 GCC28680.1 ADMH02001312 ETN63043.1 JW865237 AFO97754.1 GACK01004988 JAA60046.1 GFAC01003406 JAT95782.1 GFAA01000557 JAU02878.1 GBBK01001662 JAC22820.1 GACK01004987 JAA60047.1 AHAT01011146 AHAT01011147 KB200049 ESP03325.1 GFDF01001257 JAV12827.1 GGFK01006107 MBW39428.1 GGFK01006127 MBW39448.1 AXCM01002097 BFAA01000014 GCB62993.1 AXCN02000007
KZ150054 PZC74341.1 BABH01018251 BABH01018252 BABH01018253 BABH01018254 AGBW02004770 OWR55377.1 RSAL01000205 RVE44369.1 GAIX01014371 JAA78189.1 GECZ01027491 JAS42278.1 GEBQ01014771 JAT25206.1 KK107106 QOIP01000002 EZA59423.1 RLU25848.1 JTDY01005670 KOB66768.1 GEZM01036586 JAV82580.1 AAZX01001200 KQ977151 KYN05292.1 GL439874 EFN66661.1 KQ971354 EFA05879.1 KQ983238 KYQ46247.1 KQ980895 KYN11743.1 KQ976509 KYM82687.1 ADTU01000325 NEVH01013275 PNF28998.1 GBYB01008556 GBYB01008560 JAG78323.1 JAG78327.1 KQ414616 KOC68448.1 KK853060 KDR11903.1 KQ769179 OAD52925.1 KQ434829 KZC07749.1 GL888609 EGI59044.1 KQ981606 KYN39577.1 BT127036 AEE61998.1 KZ288369 PBC26779.1 LBMM01000238 KMR03323.1 GBHO01020351 GBHO01020349 GBHO01020348 GBRD01015144 GBRD01015143 GBRD01015142 GDHC01017288 JAG23253.1 JAG23255.1 JAG23256.1 JAG50682.1 JAQ01341.1 GBBI01003677 JAC15035.1 JH818095 EKC37504.1 GFTR01007108 JAW09318.1 PZQS01000013 PVD19594.1 GGMS01008646 MBY77849.1 RQTK01000184 RUS85048.1 KQ419065 KOF84726.1 ABLF02014212 GAPW01000800 JAC12798.1 GGMR01013839 MBY26458.1 JH432114 KK120653 KFM78747.1 GDIQ01054315 JAN40422.1 CH477376 EAT42305.1 GEGO01001012 JAR94392.1 GEFM01004520 JAP71276.1 GG666497 EEN62263.1 GDIQ01195163 JAK56562.1 GDIP01233283 LRGB01002568 JAI90118.1 KZS07100.1 GDIQ01233391 JAK18334.1 GGFJ01003958 MBW53099.1 GDIP01093053 JAM10662.1 GAMC01002950 JAC03606.1 APCN01000326 AAAB01008859 EAA07525.4 GFDL01010382 JAV24663.1 GFDL01010162 JAV24883.1 GEFH01004831 JAP63750.1 DS231862 EDS39733.1 ATLV01010990 KE524630 KFB35822.1 GANO01000284 JAB59587.1 GL732523 EFX90447.1 GGFM01003686 MBW24437.1 GEDV01008893 JAP79664.1 GBBL01000987 JAC26333.1 BEZZ01000215 GCC28680.1 ADMH02001312 ETN63043.1 JW865237 AFO97754.1 GACK01004988 JAA60046.1 GFAC01003406 JAT95782.1 GFAA01000557 JAU02878.1 GBBK01001662 JAC22820.1 GACK01004987 JAA60047.1 AHAT01011146 AHAT01011147 KB200049 ESP03325.1 GFDF01001257 JAV12827.1 GGFK01006107 MBW39428.1 GGFK01006127 MBW39448.1 AXCM01002097 BFAA01000014 GCB62993.1 AXCN02000007
Proteomes
UP000053268
UP000053240
UP000005204
UP000007151
UP000283053
UP000053097
+ More
UP000279307 UP000037510 UP000002358 UP000078542 UP000000311 UP000007266 UP000075809 UP000078492 UP000078540 UP000005205 UP000235965 UP000053825 UP000027135 UP000192223 UP000005203 UP000076502 UP000007755 UP000078541 UP000242457 UP000036403 UP000005408 UP000245119 UP000271974 UP000053454 UP000007819 UP000054359 UP000008820 UP000001554 UP000076858 UP000069272 UP000076408 UP000076407 UP000075903 UP000075840 UP000007062 UP000075902 UP000002320 UP000030765 UP000000305 UP000287033 UP000000673 UP000075882 UP000261520 UP000018468 UP000030746 UP000075883 UP000075884 UP000288216 UP000075886 UP000075881 UP000075900 UP000075920
UP000279307 UP000037510 UP000002358 UP000078542 UP000000311 UP000007266 UP000075809 UP000078492 UP000078540 UP000005205 UP000235965 UP000053825 UP000027135 UP000192223 UP000005203 UP000076502 UP000007755 UP000078541 UP000242457 UP000036403 UP000005408 UP000245119 UP000271974 UP000053454 UP000007819 UP000054359 UP000008820 UP000001554 UP000076858 UP000069272 UP000076408 UP000076407 UP000075903 UP000075840 UP000007062 UP000075902 UP000002320 UP000030765 UP000000305 UP000287033 UP000000673 UP000075882 UP000261520 UP000018468 UP000030746 UP000075883 UP000075884 UP000288216 UP000075886 UP000075881 UP000075900 UP000075920
PRIDE
Interpro
SUPFAM
SSF53383
SSF53383
Gene 3D
ProteinModelPortal
A0A2H1W3D9
A0A194QEX6
A0A194QVM3
A0A2W1BJ60
H9IX64
A0A212FNS3
+ More
A0A437B1X3 S4NMA1 A0A1B6EWH8 A0A1B6LNK9 A0A026WTX8 A0A0L7KU27 A0A1Y1MDA6 K7IV63 A0A151IL08 E2AIS2 D6WSR0 A0A151WEL7 A0A151IVJ8 A0A195BES7 A0A158NNP9 A0A2J7QK95 A0A0C9RN96 A0A0L7RC92 A0A067R2G1 A0A310SF45 A0A1W4XB13 A0A087ZYD4 A0A154P742 F4X363 A0A195FH69 J3JUX1 A0A2A3E6I6 A0A0J7P331 A0A0A9XWJ6 A0A023F0P4 K1R1W9 A0A224XHN6 A0A2T7NEJ3 A0A2S2QJB6 A0A433TU00 A0A0L8H611 J9JX24 A0A023ETX0 A0A2S2PB97 T1JL86 A0A087UN08 A0A0P6F1L9 Q177H2 A0A147BV28 A0A131XZW9 C3YBA6 A0A0N8BB37 A0A0P4Y5M3 A0A0P5H2V8 A0A182FNY0 A0A2M4BJ54 A0A0P5VZE4 A0A182YHR2 A0A182XAW1 A0A182V7H0 W8BQR4 A0A182HSR1 Q7QCB7 A0A1Q3FAZ1 A0A1Q3FBK8 A0A131XCD4 A0A182U0M6 B0W936 A0A084VCX8 U5EYI1 E9FRQ1 A0A2M3Z7E3 A0A131YK52 A0A023FZZ2 A0A401SEA9 W5JIC9 V9KIE2 L7M6Z4 A0A182LET4 A0A1E1X949 A0A1E1XUB3 A0A023FPQ0 L7M878 A0A3B3ZSQ8 W5M914 V4CLX7 A0A1L8E295 A0A2M4AFL0 A0A2M4AFM9 A0A182M8Z5 A0A182NRM6 A0A401NQ32 A0A182R055 A0A182JYH5 A0A182RS58 A0A182VWV8
A0A437B1X3 S4NMA1 A0A1B6EWH8 A0A1B6LNK9 A0A026WTX8 A0A0L7KU27 A0A1Y1MDA6 K7IV63 A0A151IL08 E2AIS2 D6WSR0 A0A151WEL7 A0A151IVJ8 A0A195BES7 A0A158NNP9 A0A2J7QK95 A0A0C9RN96 A0A0L7RC92 A0A067R2G1 A0A310SF45 A0A1W4XB13 A0A087ZYD4 A0A154P742 F4X363 A0A195FH69 J3JUX1 A0A2A3E6I6 A0A0J7P331 A0A0A9XWJ6 A0A023F0P4 K1R1W9 A0A224XHN6 A0A2T7NEJ3 A0A2S2QJB6 A0A433TU00 A0A0L8H611 J9JX24 A0A023ETX0 A0A2S2PB97 T1JL86 A0A087UN08 A0A0P6F1L9 Q177H2 A0A147BV28 A0A131XZW9 C3YBA6 A0A0N8BB37 A0A0P4Y5M3 A0A0P5H2V8 A0A182FNY0 A0A2M4BJ54 A0A0P5VZE4 A0A182YHR2 A0A182XAW1 A0A182V7H0 W8BQR4 A0A182HSR1 Q7QCB7 A0A1Q3FAZ1 A0A1Q3FBK8 A0A131XCD4 A0A182U0M6 B0W936 A0A084VCX8 U5EYI1 E9FRQ1 A0A2M3Z7E3 A0A131YK52 A0A023FZZ2 A0A401SEA9 W5JIC9 V9KIE2 L7M6Z4 A0A182LET4 A0A1E1X949 A0A1E1XUB3 A0A023FPQ0 L7M878 A0A3B3ZSQ8 W5M914 V4CLX7 A0A1L8E295 A0A2M4AFL0 A0A2M4AFM9 A0A182M8Z5 A0A182NRM6 A0A401NQ32 A0A182R055 A0A182JYH5 A0A182RS58 A0A182VWV8
Ontologies
GO
PANTHER
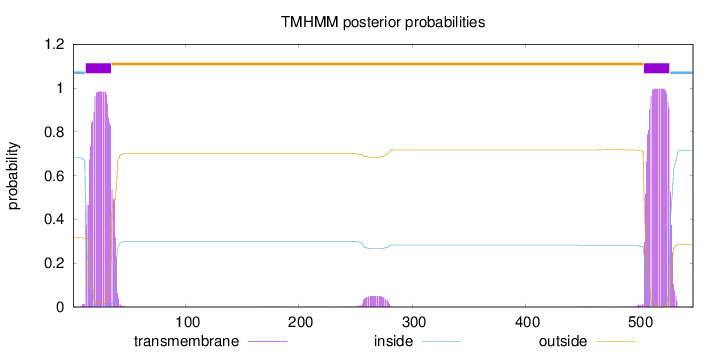
Topology
Length:
548
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
45.0801899999999
Exp number, first 60 AAs:
21.79434
Total prob of N-in:
0.68378
POSSIBLE N-term signal
sequence
inside
1 - 11
TMhelix
12 - 34
outside
35 - 504
TMhelix
505 - 527
inside
528 - 548
Population Genetic Test Statistics
Pi
32.13905
Theta
220.720345
Tajima's D
-2.203019
CLR
0.075476
CSRT
0.00659967001649917
Interpretation
Uncertain
