Gene
KWMTBOMO11320
Pre Gene Modal
BGIBMGA001982
Annotation
PREDICTED:_peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine_amidase_[Bombyx_mori]
Full name
Peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase
Alternative Name
Peptide:N-glycanase
N-glycanase 1
N-glycanase 1
Location in the cell
Nuclear Reliability : 2.028
Sequence
CDS
ATGGAGGACACAGTGCATTTAGCTTTAGTCGAGCAGAGTATTCGGAACACAGATAAATTCATTATAGCTCTCTACGAACTCTTGGAACGCATCACTCGTATTCTGGAGAATCCGCATGATTACGAATTGAGGAGTATTAAAAAAAATGTTTTCAAAGATTTATCGAAACTGGACTCTTTTAATGAATACATGAAGTATATCGGATTCAAATCGGTAGACAACGAATTTACATATCCAAAGGAGTTAAGTTTTAGCAAATTAAGGATGGCTCAAGTGGCCATCGAAAGGAAACTACATTTTTGCTGTGGATCCGTGCCTATTAGGCCGATCCCAGTAAATTCTACAGACGTTAGAGAACAACCTAAAGCATCCCCAGTTCATTCATTGCAAACTAAAAACAGATTCCTACTCAAAATTCAAGATCTATTCAATGGTATGCAAGTTTATGAGGATGAAGATTTATTAGCACATGCCCGAGACCAAATACCCTTAGTGACATTGCAGCTTATGGCTTTAGACCGAGTTAGGGAACAACAGAAGAAAATTAAGATGGGGGAAATAAAAGCAAACGATCTACCGTTTGATACAGCTTTGCTAATGGAGCTCCTGGACTGGTTCAAGCATGATTTCTTTAAATGGGTCGACAAGCCTGACTGTGATTTATGCGGAGAACGAACCGTCAATCATGAAAACGCTATCATGACGATAGAAGGTGAAACTTGTAGAGTTGAGCTCTACCAGTGCACCGTATGTGAAGGAGGAACCGCGATGTTCCCGCGCTACAATAACCTGCGTACATTGCTGCGCACGAGGAGTGGTCGGTGCGGCGAATGGGCCAATTGCTTTACGCTGCTGTGTCGTGCTTTGGGCTACGACACCAGATACGTTTATGACGTCACCGATCACGTGTGGTGCGAGGTGTTCGACTACGACTCGAACCAGTGGCTGCACGTGGACCCGTGCGAGGCGAAGCTGAACACGCCCCTCATGTACTCGCACGGCTGGGGCAAACGCCTCTCTTACGTCATCGCCTTCTCGAGGGACGACCTGCAGGACGTGACGTGGCGGTACACCACCAACCATAAAGAGGTGTTGAAATACCGCTACCTGTGCACGGAAACAGAATTGATCTCCACGATAATGACGCTGCGGAAGCACAAACAGCGCGATGTAACGGAGGCCAGGCGACGCTATCTAGCCAAGCGCACTCTCGAGGAACTAGTGCAGATGATGGTGGAAAGGAAACCGAGCGACTACGAGTCCCACGGGCGTATATCCGGGTCGCGCAAGTGGCGGATGCAGCGCGGGGAGCTGGGCGGTGCGGCGGGCGGGGCGGTGGGGGGGCACACGTTCGTGCTGGGGCGCGCGGGCACGCACGTGATGCGCTACTACTGCGGCGCCGACCGGTGGAGGCGGGCGTGCGACGGCGCCGAGACCGCGCCCCTGCCCGGCTGGCACGCGGCCGTGCTGCACGCCGCCAACGTGTTCAGGAAAGTCGAGCCGGACTGGCTGCAGTCCTACATAGCTAGAGAAGAGGGCGAGGACTTCGGTTCGATCTCGTGGGCGTTCGCGGCTAGTGAGGAGCTGACGTGCGCCAGTTTATCGATAAAGGTCCGCACGGCGCTGTACGAGAGCGGCCGCATCGACTGGACGGTGAAGTTCGACGATGAAAACCCTACCACGGTCACGTTGAGCGATAAACCAACGAAGTTCGCACGAAAGTTTCGGAAGGTCATCATCAAAGCCGAGCTGTCGGGCGGCGACGGCCCCGTGCGCTGGCAACACGCCCAGCTGTTCCGGCAACACACGTACTCCAAGAGAAGCTCCTTTATCGTCACAATCGTGGTCCAGTAA
Protein
MEDTVHLALVEQSIRNTDKFIIALYELLERITRILENPHDYELRSIKKNVFKDLSKLDSFNEYMKYIGFKSVDNEFTYPKELSFSKLRMAQVAIERKLHFCCGSVPIRPIPVNSTDVREQPKASPVHSLQTKNRFLLKIQDLFNGMQVYEDEDLLAHARDQIPLVTLQLMALDRVREQQKKIKMGEIKANDLPFDTALLMELLDWFKHDFFKWVDKPDCDLCGERTVNHENAIMTIEGETCRVELYQCTVCEGGTAMFPRYNNLRTLLRTRSGRCGEWANCFTLLCRALGYDTRYVYDVTDHVWCEVFDYDSNQWLHVDPCEAKLNTPLMYSHGWGKRLSYVIAFSRDDLQDVTWRYTTNHKEVLKYRYLCTETELISTIMTLRKHKQRDVTEARRRYLAKRTLEELVQMMVERKPSDYESHGRISGSRKWRMQRGELGGAAGGAVGGHTFVLGRAGTHVMRYYCGADRWRRACDGAETAPLPGWHAAVLHAANVFRKVEPDWLQSYIAREEGEDFGSISWAFAASEELTCASLSIKVRTALYESGRIDWTVKFDDENPTTVTLSDKPTKFARKFRKVIIKAELSGGDGPVRWQHAQLFRQHTYSKRSSFIVTIVVQ
Summary
Description
Specifically deglycosylates the denatured form of N-linked glycoproteins in the cytoplasm and assists their proteasome-mediated degradation. Cleaves the beta-aspartyl-glucosamine (GlcNAc) of the glycan and the amide side chain of Asn, converting Asn to Asp. Prefers proteins containing high-mannose over those bearing complex type oligosaccharides. Can recognize misfolded proteins in the endoplasmic reticulum that are exported to the cytosol to be destroyed and deglycosylate them, while it has no activity toward native proteins. Deglycosylation is a prerequisite for subsequent proteasome-mediated degradation of some, but not all, misfolded glycoproteins (By similarity).
Catalytic Activity
Hydrolysis of an N(4)-(acetyl-beta-D-glucosaminyl)asparagine residue in which the glucosamine residue may be further glycosylated, to yield a (substituted) N-acetyl-beta-D-glucosaminylamine and a peptide containing an aspartate residue.
Cofactor
Zn(2+)
Similarity
Belongs to the serpin family.
Belongs to the transglutaminase-like superfamily. PNGase family.
Belongs to the transglutaminase-like superfamily. PNGase family.
Keywords
Complete proteome
Cytoplasm
Hydrolase
Metal-binding
Phosphoprotein
Reference proteome
Zinc
Feature
chain Peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase
Uniprot
H9IXJ9
A0A2A4JM22
A0A212F0C5
A0A2H1W1F1
A0A2W1BNP0
A0A1E1WPZ4
+ More
A0A0L7L9N5 A0A194QX43 A0A194QEW6 A0A1Y1M5T0 A0A1Y1M1G6 A0A139WEQ5 A0A1W4WH01 A0A067QFL0 A0A1Q3FIL6 B0WT33 A0A1L8DU46 V5GPK1 B4MYN5 B4KSC7 A0A2J7R3K4 A0A0M4E457 A0A1S4G2F7 A0A2J7R3J7 Q16G66 B4MDY1 A0A2J7R3M4 Q7KRR5 Q28YQ7 A0A1I8Q858 A0A1J1INN8 B3N3G8 B4GIZ8 B4II25 B4QCQ6 A0A093GVI2 A0A3B0JXM6 A0A182GLV7 B4J9V1 B4NYW0 A0A1B0CRQ6 W8APL2 A0A3P9PDU0 A0A087XNK1 A0A3B3VUN1 G3WDQ9 A0A3B3WEZ1 A0A3Q2PTR9 U5EUG5 G3WDQ8 A0A3P9PDR4 H9GEH0 M4AR69 A0A3B3WEH7 H2SWE9 A0A1W4V2C7 A0A1A9YE51 F4WHB9 F7D099 A0A3P9KYM2 A0A034V3S8 H2LXI6 A0A0K8VS08 A0A315VXC2 T1IJ72 A0A1A8ITM1 A0A1A9V9X0 A0A2R5LEY2 A0A0L7R362 A0A0K8TMU0 A0A146ZNH6 A0A1S3JJE2 A0A3N0YI91 A0A1W4UNC6 A0A1I8MV56 A0A2K6EN32 A0A1S3K0I9 A0A158NB52 A0A3P9H9I3 G3NZ69 A0A146V9E4 A0A093QPG4 H0WKM6 A0A369SB05 A0A195D6P1 B3MJ81 B3RRV6 A0A1Z5KUR9 A0A099YVZ1 A0A0L0CPF8 A0A0F8C8M8 A0A3Q3VZD1 A0A224Z3V3 W5UQD7 A7SSF4 A0A131XD48 A0A091FNN0 Q5ZJM3 F7AI61 A0A093KCM9
A0A0L7L9N5 A0A194QX43 A0A194QEW6 A0A1Y1M5T0 A0A1Y1M1G6 A0A139WEQ5 A0A1W4WH01 A0A067QFL0 A0A1Q3FIL6 B0WT33 A0A1L8DU46 V5GPK1 B4MYN5 B4KSC7 A0A2J7R3K4 A0A0M4E457 A0A1S4G2F7 A0A2J7R3J7 Q16G66 B4MDY1 A0A2J7R3M4 Q7KRR5 Q28YQ7 A0A1I8Q858 A0A1J1INN8 B3N3G8 B4GIZ8 B4II25 B4QCQ6 A0A093GVI2 A0A3B0JXM6 A0A182GLV7 B4J9V1 B4NYW0 A0A1B0CRQ6 W8APL2 A0A3P9PDU0 A0A087XNK1 A0A3B3VUN1 G3WDQ9 A0A3B3WEZ1 A0A3Q2PTR9 U5EUG5 G3WDQ8 A0A3P9PDR4 H9GEH0 M4AR69 A0A3B3WEH7 H2SWE9 A0A1W4V2C7 A0A1A9YE51 F4WHB9 F7D099 A0A3P9KYM2 A0A034V3S8 H2LXI6 A0A0K8VS08 A0A315VXC2 T1IJ72 A0A1A8ITM1 A0A1A9V9X0 A0A2R5LEY2 A0A0L7R362 A0A0K8TMU0 A0A146ZNH6 A0A1S3JJE2 A0A3N0YI91 A0A1W4UNC6 A0A1I8MV56 A0A2K6EN32 A0A1S3K0I9 A0A158NB52 A0A3P9H9I3 G3NZ69 A0A146V9E4 A0A093QPG4 H0WKM6 A0A369SB05 A0A195D6P1 B3MJ81 B3RRV6 A0A1Z5KUR9 A0A099YVZ1 A0A0L0CPF8 A0A0F8C8M8 A0A3Q3VZD1 A0A224Z3V3 W5UQD7 A7SSF4 A0A131XD48 A0A091FNN0 Q5ZJM3 F7AI61 A0A093KCM9
EC Number
3.5.1.52
Pubmed
19121390
22118469
28756777
26227816
26354079
28004739
+ More
18362917 19820115 24845553 17994087 17510324 18057021 10831608 10731132 12537572 12537569 18327897 15632085 22936249 26483478 17550304 24495485 21709235 21881562 23542700 21551351 21719571 17495919 17554307 25348373 29703783 26369729 26383154 25315136 21347285 30042472 18719581 28528879 26108605 25835551 28797301 23127152 17615350 28049606 15642098
18362917 19820115 24845553 17994087 17510324 18057021 10831608 10731132 12537572 12537569 18327897 15632085 22936249 26483478 17550304 24495485 21709235 21881562 23542700 21551351 21719571 17495919 17554307 25348373 29703783 26369729 26383154 25315136 21347285 30042472 18719581 28528879 26108605 25835551 28797301 23127152 17615350 28049606 15642098
EMBL
BABH01018274
BABH01018275
NWSH01001016
PCG73107.1
AGBW02011140
OWR47151.1
+ More
ODYU01005752 SOQ46925.1 KZ150054 PZC74350.1 GDQN01002002 JAT89052.1 JTDY01002074 KOB72172.1 KQ461073 KPJ09530.1 KQ459053 KPJ04083.1 GEZM01043172 JAV79256.1 GEZM01043176 JAV79251.1 KQ971354 KYB26414.1 KK853579 KDR06555.1 GFDL01007615 JAV27430.1 DS232080 EDS34176.1 GFDF01004121 JAV09963.1 GALX01002422 JAB66044.1 CH963894 EDW77224.1 CH933808 EDW08409.1 NEVH01007819 PNF35419.1 CP012524 ALC40741.1 PNF35421.1 CH478322 EAT33229.1 CH940662 EDW58746.1 KRF78452.1 PNF35420.1 AF250926 AE013599 AY119239 BT001557 CM000071 CVRI01000054 CRL00097.1 CH954177 EDV59850.1 CH479183 EDW36416.1 CH480841 EDW49551.1 CM000362 CM002911 EDX05845.1 KMY91682.1 KL216727 KFV71082.1 OUUW01000001 SPP75838.1 JXUM01073164 JXUM01073165 KQ562759 KXJ75179.1 CH916367 EDW01515.1 CM000157 EDW89811.1 AJWK01025119 AJWK01025120 GAMC01018588 JAB87967.1 AYCK01007384 AEFK01202071 AEFK01202072 AEFK01202073 AEFK01202074 GANO01001483 JAB58388.1 AAWZ02026560 GL888158 EGI66396.1 GAKP01022529 GAKP01022527 JAC36425.1 GDHF01010652 JAI41662.1 NHOQ01001491 PWA24111.1 JH430232 HAED01014246 SBR00691.1 GGLE01003882 MBY08008.1 KQ414663 KOC65312.1 GDAI01001944 JAI15659.1 GCES01018575 JAR67748.1 RJVU01042534 ROL45873.1 ADTU01010884 GCES01072341 JAR13982.1 KL426509 KFW90446.1 AAQR03005117 AAQR03005118 AAQR03005119 AAQR03005120 AAQR03005121 AAQR03005122 NOWV01000034 RDD43711.1 KQ976760 KYN08531.1 CH902619 EDV38175.1 DS985243 EDV26937.1 GFJQ02008127 JAV98842.1 KL886993 KGL74389.1 JRES01000204 KNC33319.1 KQ041387 KKF27325.1 GFPF01010455 MAA21601.1 JT415651 AHH41778.1 DS469776 EDO33371.1 GEFH01005080 JAP63501.1 KL447195 KFO70734.1 AJ720411 KL206987 KFV88029.1
ODYU01005752 SOQ46925.1 KZ150054 PZC74350.1 GDQN01002002 JAT89052.1 JTDY01002074 KOB72172.1 KQ461073 KPJ09530.1 KQ459053 KPJ04083.1 GEZM01043172 JAV79256.1 GEZM01043176 JAV79251.1 KQ971354 KYB26414.1 KK853579 KDR06555.1 GFDL01007615 JAV27430.1 DS232080 EDS34176.1 GFDF01004121 JAV09963.1 GALX01002422 JAB66044.1 CH963894 EDW77224.1 CH933808 EDW08409.1 NEVH01007819 PNF35419.1 CP012524 ALC40741.1 PNF35421.1 CH478322 EAT33229.1 CH940662 EDW58746.1 KRF78452.1 PNF35420.1 AF250926 AE013599 AY119239 BT001557 CM000071 CVRI01000054 CRL00097.1 CH954177 EDV59850.1 CH479183 EDW36416.1 CH480841 EDW49551.1 CM000362 CM002911 EDX05845.1 KMY91682.1 KL216727 KFV71082.1 OUUW01000001 SPP75838.1 JXUM01073164 JXUM01073165 KQ562759 KXJ75179.1 CH916367 EDW01515.1 CM000157 EDW89811.1 AJWK01025119 AJWK01025120 GAMC01018588 JAB87967.1 AYCK01007384 AEFK01202071 AEFK01202072 AEFK01202073 AEFK01202074 GANO01001483 JAB58388.1 AAWZ02026560 GL888158 EGI66396.1 GAKP01022529 GAKP01022527 JAC36425.1 GDHF01010652 JAI41662.1 NHOQ01001491 PWA24111.1 JH430232 HAED01014246 SBR00691.1 GGLE01003882 MBY08008.1 KQ414663 KOC65312.1 GDAI01001944 JAI15659.1 GCES01018575 JAR67748.1 RJVU01042534 ROL45873.1 ADTU01010884 GCES01072341 JAR13982.1 KL426509 KFW90446.1 AAQR03005117 AAQR03005118 AAQR03005119 AAQR03005120 AAQR03005121 AAQR03005122 NOWV01000034 RDD43711.1 KQ976760 KYN08531.1 CH902619 EDV38175.1 DS985243 EDV26937.1 GFJQ02008127 JAV98842.1 KL886993 KGL74389.1 JRES01000204 KNC33319.1 KQ041387 KKF27325.1 GFPF01010455 MAA21601.1 JT415651 AHH41778.1 DS469776 EDO33371.1 GEFH01005080 JAP63501.1 KL447195 KFO70734.1 AJ720411 KL206987 KFV88029.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000037510
UP000053240
UP000053268
+ More
UP000007266 UP000192223 UP000027135 UP000002320 UP000007798 UP000009192 UP000235965 UP000092553 UP000008820 UP000008792 UP000000803 UP000001819 UP000095300 UP000183832 UP000008711 UP000008744 UP000001292 UP000000304 UP000053875 UP000268350 UP000069940 UP000249989 UP000001070 UP000002282 UP000092461 UP000242638 UP000028760 UP000261500 UP000007648 UP000261480 UP000265000 UP000001646 UP000002852 UP000005226 UP000192221 UP000092443 UP000007755 UP000002280 UP000265180 UP000001038 UP000078200 UP000053825 UP000085678 UP000095301 UP000233160 UP000005205 UP000265200 UP000007635 UP000005225 UP000253843 UP000078542 UP000007801 UP000009022 UP000053641 UP000037069 UP000261620 UP000221080 UP000001593 UP000053760 UP000000539 UP000008225 UP000053584
UP000007266 UP000192223 UP000027135 UP000002320 UP000007798 UP000009192 UP000235965 UP000092553 UP000008820 UP000008792 UP000000803 UP000001819 UP000095300 UP000183832 UP000008711 UP000008744 UP000001292 UP000000304 UP000053875 UP000268350 UP000069940 UP000249989 UP000001070 UP000002282 UP000092461 UP000242638 UP000028760 UP000261500 UP000007648 UP000261480 UP000265000 UP000001646 UP000002852 UP000005226 UP000192221 UP000092443 UP000007755 UP000002280 UP000265180 UP000001038 UP000078200 UP000053825 UP000085678 UP000095301 UP000233160 UP000005205 UP000265200 UP000007635 UP000005225 UP000253843 UP000078542 UP000007801 UP000009022 UP000053641 UP000037069 UP000261620 UP000221080 UP000001593 UP000053760 UP000000539 UP000008225 UP000053584
Interpro
IPR002931
Transglutaminase-like
+ More
IPR008979 Galactose-bd-like_sf
IPR038680 PAW_sf
IPR038765 Papain-like_cys_pep_sf
IPR006588 Peptide_N_glycanase_PAW_dom
IPR036339 PUB-like_dom_sf
IPR018997 PUB_domain
IPR036186 Serpin_sf
IPR042178 Serpin_sf_1
IPR000215 Serpin_fam
IPR023796 Serpin_dom
IPR023795 Serpin_CS
IPR042185 Serpin_sf_2
IPR018325 Rad4/PNGase_transGLS-fold
IPR008979 Galactose-bd-like_sf
IPR038680 PAW_sf
IPR038765 Papain-like_cys_pep_sf
IPR006588 Peptide_N_glycanase_PAW_dom
IPR036339 PUB-like_dom_sf
IPR018997 PUB_domain
IPR036186 Serpin_sf
IPR042178 Serpin_sf_1
IPR000215 Serpin_fam
IPR023796 Serpin_dom
IPR023795 Serpin_CS
IPR042185 Serpin_sf_2
IPR018325 Rad4/PNGase_transGLS-fold
Gene 3D
ProteinModelPortal
H9IXJ9
A0A2A4JM22
A0A212F0C5
A0A2H1W1F1
A0A2W1BNP0
A0A1E1WPZ4
+ More
A0A0L7L9N5 A0A194QX43 A0A194QEW6 A0A1Y1M5T0 A0A1Y1M1G6 A0A139WEQ5 A0A1W4WH01 A0A067QFL0 A0A1Q3FIL6 B0WT33 A0A1L8DU46 V5GPK1 B4MYN5 B4KSC7 A0A2J7R3K4 A0A0M4E457 A0A1S4G2F7 A0A2J7R3J7 Q16G66 B4MDY1 A0A2J7R3M4 Q7KRR5 Q28YQ7 A0A1I8Q858 A0A1J1INN8 B3N3G8 B4GIZ8 B4II25 B4QCQ6 A0A093GVI2 A0A3B0JXM6 A0A182GLV7 B4J9V1 B4NYW0 A0A1B0CRQ6 W8APL2 A0A3P9PDU0 A0A087XNK1 A0A3B3VUN1 G3WDQ9 A0A3B3WEZ1 A0A3Q2PTR9 U5EUG5 G3WDQ8 A0A3P9PDR4 H9GEH0 M4AR69 A0A3B3WEH7 H2SWE9 A0A1W4V2C7 A0A1A9YE51 F4WHB9 F7D099 A0A3P9KYM2 A0A034V3S8 H2LXI6 A0A0K8VS08 A0A315VXC2 T1IJ72 A0A1A8ITM1 A0A1A9V9X0 A0A2R5LEY2 A0A0L7R362 A0A0K8TMU0 A0A146ZNH6 A0A1S3JJE2 A0A3N0YI91 A0A1W4UNC6 A0A1I8MV56 A0A2K6EN32 A0A1S3K0I9 A0A158NB52 A0A3P9H9I3 G3NZ69 A0A146V9E4 A0A093QPG4 H0WKM6 A0A369SB05 A0A195D6P1 B3MJ81 B3RRV6 A0A1Z5KUR9 A0A099YVZ1 A0A0L0CPF8 A0A0F8C8M8 A0A3Q3VZD1 A0A224Z3V3 W5UQD7 A7SSF4 A0A131XD48 A0A091FNN0 Q5ZJM3 F7AI61 A0A093KCM9
A0A0L7L9N5 A0A194QX43 A0A194QEW6 A0A1Y1M5T0 A0A1Y1M1G6 A0A139WEQ5 A0A1W4WH01 A0A067QFL0 A0A1Q3FIL6 B0WT33 A0A1L8DU46 V5GPK1 B4MYN5 B4KSC7 A0A2J7R3K4 A0A0M4E457 A0A1S4G2F7 A0A2J7R3J7 Q16G66 B4MDY1 A0A2J7R3M4 Q7KRR5 Q28YQ7 A0A1I8Q858 A0A1J1INN8 B3N3G8 B4GIZ8 B4II25 B4QCQ6 A0A093GVI2 A0A3B0JXM6 A0A182GLV7 B4J9V1 B4NYW0 A0A1B0CRQ6 W8APL2 A0A3P9PDU0 A0A087XNK1 A0A3B3VUN1 G3WDQ9 A0A3B3WEZ1 A0A3Q2PTR9 U5EUG5 G3WDQ8 A0A3P9PDR4 H9GEH0 M4AR69 A0A3B3WEH7 H2SWE9 A0A1W4V2C7 A0A1A9YE51 F4WHB9 F7D099 A0A3P9KYM2 A0A034V3S8 H2LXI6 A0A0K8VS08 A0A315VXC2 T1IJ72 A0A1A8ITM1 A0A1A9V9X0 A0A2R5LEY2 A0A0L7R362 A0A0K8TMU0 A0A146ZNH6 A0A1S3JJE2 A0A3N0YI91 A0A1W4UNC6 A0A1I8MV56 A0A2K6EN32 A0A1S3K0I9 A0A158NB52 A0A3P9H9I3 G3NZ69 A0A146V9E4 A0A093QPG4 H0WKM6 A0A369SB05 A0A195D6P1 B3MJ81 B3RRV6 A0A1Z5KUR9 A0A099YVZ1 A0A0L0CPF8 A0A0F8C8M8 A0A3Q3VZD1 A0A224Z3V3 W5UQD7 A7SSF4 A0A131XD48 A0A091FNN0 Q5ZJM3 F7AI61 A0A093KCM9
PDB
2F4O
E-value=1.58346e-51,
Score=514
Ontologies
GO
PANTHER
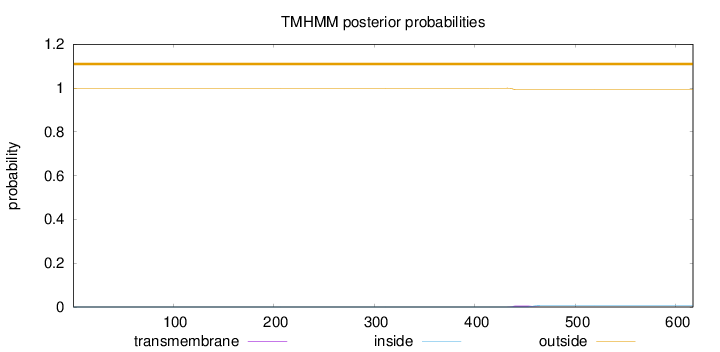
Topology
Subcellular location
Cytoplasm
Length:
617
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.13835
Exp number, first 60 AAs:
0.00015
Total prob of N-in:
0.00048
outside
1 - 617
Population Genetic Test Statistics
Pi
216.875494
Theta
156.580092
Tajima's D
1.774806
CLR
0.350544
CSRT
0.848307584620769
Interpretation
Uncertain
