Pre Gene Modal
BGIBMGA001984
Annotation
PREDICTED:_putative_deoxyribonuclease_TATDN1_[Plutella_xylostella]
Location in the cell
Nuclear Reliability : 2.81
Sequence
CDS
ATGAGTGCGTTAAGAAAATATATAGACATTGGTGCTAACCTAACAGATGATATGTATAAAGGAGAATATAATGGGTCTAGAAAACATGATCCAGATTTGGACGTGGTACTAGAGAGAGCTTGGACTAGTGGCATCAACAAAATTATTGTTACTGGAGGGAGTCTTGAAGACAGTAAAAAAGCAATCGAGCTGTCTAGGACAGATTCTAAGCTATTCACAACTGTGGGATGTCATCCAACCAGGTGCAGTGAGTTTCTATCCAATCCTGATGACTACCTACAAGGACTGCGAGATCTCATATCAGGAAACAAGGACAAAGTTGTGGCTATAGGAGAGTGTGGACTAGATTATGAAAGGTTTCACTTTTGTGAGAAGGAAGTGCAACTAAAGTAG
Protein
MSALRKYIDIGANLTDDMYKGEYNGSRKHDPDLDVVLERAWTSGINKIIVTGGSLEDSKKAIELSRTDSKLFTTVGCHPTRCSEFLSNPDDYLQGLRDLISGNKDKVVAIGECGLDYERFHFCEKEVQLK
Summary
Uniprot
H9IXK1
A0A2W1BRH2
A0A194QGL7
A0A2H1VER8
A0A3S2NNI6
A0A2A3EQZ1
+ More
A0A087ZUX7 A0A154PP52 A0A158NNV7 A0A2J7PI09 A0A3L8DN63 S4PV16 R4WJX6 A0A1B6I729 A0A1B6ILS4 A0A151IC92 E2A8B2 A0A1B6KU28 A0A1W4XHM8 A0A151IWT8 J3JXH3 N6SRK1 U4U2X9 A0A212F099 A0A1B6JIE8 A0A2R7VPJ1 A0A1Y1LYK0 A0A151WJT5 A0A1B6FIY3 A0A0L7RAN1 A0A2P8Z375 A0A182YKU1 A0A182RDL3 B4KQJ9 A0A1B0C7X8 A0A1A9YGJ7 B3MGQ9 A0A182WLL5 A0A1B6CB08 A0A195BTN0 A0A1I8MXX4 A0A182SLY9 D6WQZ9 A0A1I8P2V7 A0A182KNX4 B4ME40 A0A0M8ZUG0 A0A0L8FYA4 D3TS77 A0A0L8FYI5 A0A182I1B7 A0A182TUJ7 A0A1Q3FIY5 A0A074ZAQ7 A0A182VMP7 Q7Q5W8 A0A182XPB1 H2KUU8 B4MJS8 W5JQZ7 B0XK00 A0A1S8WHN6 A0A0K8WBY6 W8C986 A0A182NK26 A0A1S3IPA1 B4QDS5 A0A232EV92 C9QP59 A0A0J9R6Y5 A0A084WEP0 A0A2H1BSP3 A0A3B0JI32 B4P1A5 A0A146M136 A0A182IVP1 A0A0K8W9G7 A0A0P4WG58 B7Q017 A0A034WR03 A0A034WS82 A0A182GUK2 A0A0T6B6F6 B4HQD8 A0A1W4V1P6 A0A0A9ZF80 A0A0L0BTX4 B3NAL1 A0A182QNR9 Q16EQ7 A0A182FH51 A0A2H8TGL3 A0A2S2Q6F1 T1ILX7 T2MBU4 C4WSN5 A0A226ETW2 A0A2M4BU96 A0A1A9W9K8 A0A3Q1JW70
A0A087ZUX7 A0A154PP52 A0A158NNV7 A0A2J7PI09 A0A3L8DN63 S4PV16 R4WJX6 A0A1B6I729 A0A1B6ILS4 A0A151IC92 E2A8B2 A0A1B6KU28 A0A1W4XHM8 A0A151IWT8 J3JXH3 N6SRK1 U4U2X9 A0A212F099 A0A1B6JIE8 A0A2R7VPJ1 A0A1Y1LYK0 A0A151WJT5 A0A1B6FIY3 A0A0L7RAN1 A0A2P8Z375 A0A182YKU1 A0A182RDL3 B4KQJ9 A0A1B0C7X8 A0A1A9YGJ7 B3MGQ9 A0A182WLL5 A0A1B6CB08 A0A195BTN0 A0A1I8MXX4 A0A182SLY9 D6WQZ9 A0A1I8P2V7 A0A182KNX4 B4ME40 A0A0M8ZUG0 A0A0L8FYA4 D3TS77 A0A0L8FYI5 A0A182I1B7 A0A182TUJ7 A0A1Q3FIY5 A0A074ZAQ7 A0A182VMP7 Q7Q5W8 A0A182XPB1 H2KUU8 B4MJS8 W5JQZ7 B0XK00 A0A1S8WHN6 A0A0K8WBY6 W8C986 A0A182NK26 A0A1S3IPA1 B4QDS5 A0A232EV92 C9QP59 A0A0J9R6Y5 A0A084WEP0 A0A2H1BSP3 A0A3B0JI32 B4P1A5 A0A146M136 A0A182IVP1 A0A0K8W9G7 A0A0P4WG58 B7Q017 A0A034WR03 A0A034WS82 A0A182GUK2 A0A0T6B6F6 B4HQD8 A0A1W4V1P6 A0A0A9ZF80 A0A0L0BTX4 B3NAL1 A0A182QNR9 Q16EQ7 A0A182FH51 A0A2H8TGL3 A0A2S2Q6F1 T1ILX7 T2MBU4 C4WSN5 A0A226ETW2 A0A2M4BU96 A0A1A9W9K8 A0A3Q1JW70
Pubmed
19121390
28756777
26354079
21347285
30249741
23622113
+ More
23691247 20798317 22516182 23537049 22118469 28004739 29403074 25244985 17994087 25315136 18362917 19820115 20966253 20353571 12364791 14747013 17210077 22023798 20920257 23761445 24495485 28648823 22936249 24438588 17550304 26823975 25348373 26483478 25401762 26108605 17510324 24065732
23691247 20798317 22516182 23537049 22118469 28004739 29403074 25244985 17994087 25315136 18362917 19820115 20966253 20353571 12364791 14747013 17210077 22023798 20920257 23761445 24495485 28648823 22936249 24438588 17550304 26823975 25348373 26483478 25401762 26108605 17510324 24065732
EMBL
BABH01018278
KZ150054
PZC74353.1
KQ459053
KPJ04080.1
ODYU01002171
+ More
SOQ39325.1 RSAL01000205 RVE44359.1 KZ288193 PBC34185.1 KQ435007 KZC13649.1 ADTU01021806 NEVH01025130 PNF15958.1 QOIP01000006 RLU21874.1 GAIX01009323 JAA83237.1 AK417920 BAN21135.1 GECU01025003 JAS82703.1 GECU01019818 JAS87888.1 KQ978062 KYM97707.1 GL437526 EFN70325.1 GEBQ01025022 JAT14955.1 KQ980843 KYN12296.1 BT127942 AEE62904.1 APGK01059415 KB741293 ENN70244.1 KB632046 ERL88254.1 AGBW02011140 OWR47152.1 GECU01008824 JAS98882.1 KK854011 PTY09286.1 GEZM01043889 JAV78684.1 KQ983031 KYQ48122.1 GECZ01019607 JAS50162.1 KQ414618 KOC67816.1 PYGN01000217 PSN50956.1 CH933808 EDW08168.2 JXJN01007873 CH902619 EDV36817.2 GEDC01026823 JAS10475.1 KQ976417 KYM89779.1 KQ971351 EFA06484.1 CH940662 EDW58805.2 KQ435845 KOX71197.1 KQ425220 KOF69732.1 EZ424279 ADD20555.1 KOF69733.1 APCN01000049 GFDL01007560 JAV27485.1 KL596820 KER24173.1 AAAB01008960 EAA11869.4 DF144198 GAA36407.2 CH963846 EDW72367.2 ADMH02000687 ETN65325.1 DS233663 EDS31031.1 KV906970 OON13970.1 GDHF01003717 JAI48597.1 GAMC01002379 JAC04177.1 CM000362 EDX05937.1 NNAY01002039 OXU22255.1 BT099941 ACX36517.1 CM002911 KMY91853.1 ATLV01023241 KE525341 KFB48684.1 KZ437690 PIS78237.1 OUUW01000001 SPP73059.1 CM000157 EDW89107.1 GDHC01006053 JAQ12576.1 GDHF01005295 GDHF01004630 JAI47019.1 JAI47684.1 GDRN01074423 JAI63246.1 ABJB010491580 ABJB010503714 DS829427 EEC12189.1 GAKP01002332 JAC56620.1 GAKP01002334 JAC56618.1 JXUM01089369 JXUM01089370 JXUM01089371 JXUM01089372 JXUM01089373 KQ563765 KXJ73348.1 LJIG01009561 KRT82833.1 CH480816 EDW46677.1 GBHO01001526 JAG42078.1 JRES01001351 KNC23461.1 CH954177 EDV59765.2 AXCN02000497 CH478615 EAT32717.1 GFXV01001429 MBW13234.1 GGMS01003977 MBY73180.1 JH430921 HAAD01003190 CDG69422.1 ABLF02033862 AK340264 BAH70905.1 LNIX01000002 OXA61055.1 GGFJ01007515 MBW56656.1
SOQ39325.1 RSAL01000205 RVE44359.1 KZ288193 PBC34185.1 KQ435007 KZC13649.1 ADTU01021806 NEVH01025130 PNF15958.1 QOIP01000006 RLU21874.1 GAIX01009323 JAA83237.1 AK417920 BAN21135.1 GECU01025003 JAS82703.1 GECU01019818 JAS87888.1 KQ978062 KYM97707.1 GL437526 EFN70325.1 GEBQ01025022 JAT14955.1 KQ980843 KYN12296.1 BT127942 AEE62904.1 APGK01059415 KB741293 ENN70244.1 KB632046 ERL88254.1 AGBW02011140 OWR47152.1 GECU01008824 JAS98882.1 KK854011 PTY09286.1 GEZM01043889 JAV78684.1 KQ983031 KYQ48122.1 GECZ01019607 JAS50162.1 KQ414618 KOC67816.1 PYGN01000217 PSN50956.1 CH933808 EDW08168.2 JXJN01007873 CH902619 EDV36817.2 GEDC01026823 JAS10475.1 KQ976417 KYM89779.1 KQ971351 EFA06484.1 CH940662 EDW58805.2 KQ435845 KOX71197.1 KQ425220 KOF69732.1 EZ424279 ADD20555.1 KOF69733.1 APCN01000049 GFDL01007560 JAV27485.1 KL596820 KER24173.1 AAAB01008960 EAA11869.4 DF144198 GAA36407.2 CH963846 EDW72367.2 ADMH02000687 ETN65325.1 DS233663 EDS31031.1 KV906970 OON13970.1 GDHF01003717 JAI48597.1 GAMC01002379 JAC04177.1 CM000362 EDX05937.1 NNAY01002039 OXU22255.1 BT099941 ACX36517.1 CM002911 KMY91853.1 ATLV01023241 KE525341 KFB48684.1 KZ437690 PIS78237.1 OUUW01000001 SPP73059.1 CM000157 EDW89107.1 GDHC01006053 JAQ12576.1 GDHF01005295 GDHF01004630 JAI47019.1 JAI47684.1 GDRN01074423 JAI63246.1 ABJB010491580 ABJB010503714 DS829427 EEC12189.1 GAKP01002332 JAC56620.1 GAKP01002334 JAC56618.1 JXUM01089369 JXUM01089370 JXUM01089371 JXUM01089372 JXUM01089373 KQ563765 KXJ73348.1 LJIG01009561 KRT82833.1 CH480816 EDW46677.1 GBHO01001526 JAG42078.1 JRES01001351 KNC23461.1 CH954177 EDV59765.2 AXCN02000497 CH478615 EAT32717.1 GFXV01001429 MBW13234.1 GGMS01003977 MBY73180.1 JH430921 HAAD01003190 CDG69422.1 ABLF02033862 AK340264 BAH70905.1 LNIX01000002 OXA61055.1 GGFJ01007515 MBW56656.1
Proteomes
UP000005204
UP000053268
UP000283053
UP000242457
UP000005203
UP000076502
+ More
UP000005205 UP000235965 UP000279307 UP000078542 UP000000311 UP000192223 UP000078492 UP000019118 UP000030742 UP000007151 UP000075809 UP000053825 UP000245037 UP000076408 UP000075900 UP000009192 UP000092460 UP000092443 UP000007801 UP000075920 UP000078540 UP000095301 UP000075901 UP000007266 UP000095300 UP000075882 UP000008792 UP000053105 UP000053454 UP000075840 UP000075902 UP000075903 UP000007062 UP000076407 UP000007798 UP000000673 UP000002320 UP000075884 UP000085678 UP000000304 UP000215335 UP000030765 UP000268350 UP000002282 UP000075880 UP000001555 UP000069940 UP000249989 UP000001292 UP000192221 UP000037069 UP000008711 UP000075886 UP000008820 UP000069272 UP000007819 UP000198287 UP000091820 UP000265040
UP000005205 UP000235965 UP000279307 UP000078542 UP000000311 UP000192223 UP000078492 UP000019118 UP000030742 UP000007151 UP000075809 UP000053825 UP000245037 UP000076408 UP000075900 UP000009192 UP000092460 UP000092443 UP000007801 UP000075920 UP000078540 UP000095301 UP000075901 UP000007266 UP000095300 UP000075882 UP000008792 UP000053105 UP000053454 UP000075840 UP000075902 UP000075903 UP000007062 UP000076407 UP000007798 UP000000673 UP000002320 UP000075884 UP000085678 UP000000304 UP000215335 UP000030765 UP000268350 UP000002282 UP000075880 UP000001555 UP000069940 UP000249989 UP000001292 UP000192221 UP000037069 UP000008711 UP000075886 UP000008820 UP000069272 UP000007819 UP000198287 UP000091820 UP000265040
PRIDE
Interpro
SUPFAM
SSF51556
SSF51556
Gene 3D
ProteinModelPortal
H9IXK1
A0A2W1BRH2
A0A194QGL7
A0A2H1VER8
A0A3S2NNI6
A0A2A3EQZ1
+ More
A0A087ZUX7 A0A154PP52 A0A158NNV7 A0A2J7PI09 A0A3L8DN63 S4PV16 R4WJX6 A0A1B6I729 A0A1B6ILS4 A0A151IC92 E2A8B2 A0A1B6KU28 A0A1W4XHM8 A0A151IWT8 J3JXH3 N6SRK1 U4U2X9 A0A212F099 A0A1B6JIE8 A0A2R7VPJ1 A0A1Y1LYK0 A0A151WJT5 A0A1B6FIY3 A0A0L7RAN1 A0A2P8Z375 A0A182YKU1 A0A182RDL3 B4KQJ9 A0A1B0C7X8 A0A1A9YGJ7 B3MGQ9 A0A182WLL5 A0A1B6CB08 A0A195BTN0 A0A1I8MXX4 A0A182SLY9 D6WQZ9 A0A1I8P2V7 A0A182KNX4 B4ME40 A0A0M8ZUG0 A0A0L8FYA4 D3TS77 A0A0L8FYI5 A0A182I1B7 A0A182TUJ7 A0A1Q3FIY5 A0A074ZAQ7 A0A182VMP7 Q7Q5W8 A0A182XPB1 H2KUU8 B4MJS8 W5JQZ7 B0XK00 A0A1S8WHN6 A0A0K8WBY6 W8C986 A0A182NK26 A0A1S3IPA1 B4QDS5 A0A232EV92 C9QP59 A0A0J9R6Y5 A0A084WEP0 A0A2H1BSP3 A0A3B0JI32 B4P1A5 A0A146M136 A0A182IVP1 A0A0K8W9G7 A0A0P4WG58 B7Q017 A0A034WR03 A0A034WS82 A0A182GUK2 A0A0T6B6F6 B4HQD8 A0A1W4V1P6 A0A0A9ZF80 A0A0L0BTX4 B3NAL1 A0A182QNR9 Q16EQ7 A0A182FH51 A0A2H8TGL3 A0A2S2Q6F1 T1ILX7 T2MBU4 C4WSN5 A0A226ETW2 A0A2M4BU96 A0A1A9W9K8 A0A3Q1JW70
A0A087ZUX7 A0A154PP52 A0A158NNV7 A0A2J7PI09 A0A3L8DN63 S4PV16 R4WJX6 A0A1B6I729 A0A1B6ILS4 A0A151IC92 E2A8B2 A0A1B6KU28 A0A1W4XHM8 A0A151IWT8 J3JXH3 N6SRK1 U4U2X9 A0A212F099 A0A1B6JIE8 A0A2R7VPJ1 A0A1Y1LYK0 A0A151WJT5 A0A1B6FIY3 A0A0L7RAN1 A0A2P8Z375 A0A182YKU1 A0A182RDL3 B4KQJ9 A0A1B0C7X8 A0A1A9YGJ7 B3MGQ9 A0A182WLL5 A0A1B6CB08 A0A195BTN0 A0A1I8MXX4 A0A182SLY9 D6WQZ9 A0A1I8P2V7 A0A182KNX4 B4ME40 A0A0M8ZUG0 A0A0L8FYA4 D3TS77 A0A0L8FYI5 A0A182I1B7 A0A182TUJ7 A0A1Q3FIY5 A0A074ZAQ7 A0A182VMP7 Q7Q5W8 A0A182XPB1 H2KUU8 B4MJS8 W5JQZ7 B0XK00 A0A1S8WHN6 A0A0K8WBY6 W8C986 A0A182NK26 A0A1S3IPA1 B4QDS5 A0A232EV92 C9QP59 A0A0J9R6Y5 A0A084WEP0 A0A2H1BSP3 A0A3B0JI32 B4P1A5 A0A146M136 A0A182IVP1 A0A0K8W9G7 A0A0P4WG58 B7Q017 A0A034WR03 A0A034WS82 A0A182GUK2 A0A0T6B6F6 B4HQD8 A0A1W4V1P6 A0A0A9ZF80 A0A0L0BTX4 B3NAL1 A0A182QNR9 Q16EQ7 A0A182FH51 A0A2H8TGL3 A0A2S2Q6F1 T1ILX7 T2MBU4 C4WSN5 A0A226ETW2 A0A2M4BU96 A0A1A9W9K8 A0A3Q1JW70
PDB
2XIO
E-value=1.00602e-36,
Score=378
Ontologies
GO
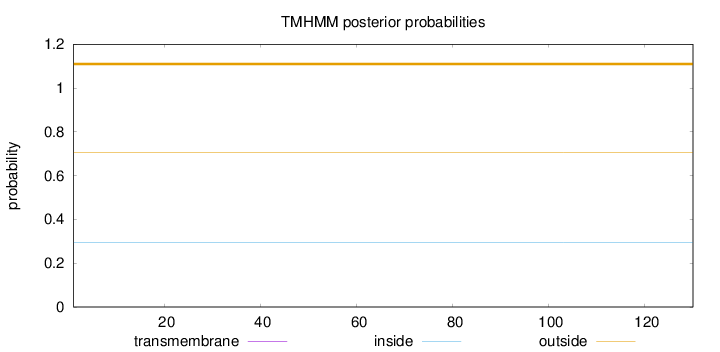
Topology
Length:
130
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00041
Exp number, first 60 AAs:
0.00041
Total prob of N-in:
0.29503
outside
1 - 130
Population Genetic Test Statistics
Pi
128.582497
Theta
76.275679
Tajima's D
1.612344
CLR
0
CSRT
0.806509674516274
Interpretation
Uncertain
