Pre Gene Modal
BGIBMGA001842
Annotation
Transmembrane_protein_214_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.042 Mitochondrial Reliability : 1.253 Nuclear Reliability : 1.541
Sequence
CDS
ATGTCTAGCGGTCAATGGGAAGTCGTAGGTAAAAATAAGAAGTCTCAGAACGGAAAGTTAAAATCCAAGGAGGATGAGAAAAAAGCTTTGAAAAATGGACCTAAACTTGAGGACGTCGTTCCTCATTCTCAAATTAAATCATTATACGCAGGAATGGATGTTGATGGCGAAAAAAAATCGGCGAAAGAGAAAAAAAACAGTGACAAGAAAGGAAAAAAACAAGAGAAGAAAGTTGAACCGCCCAAACCAAAAATCCCTAAAACCATTGAAGGTGCTCTAGAAGCCATTGACGTAACCGAGCTGGCCAGTATCATTACATCGAATAAGGTACGCTTCTCAAATGCGCCCTTAGTCTGGCTTAAGGAAGTCGCAAACTTTTTGAACAGCAAAATTCCAATTGAAGTTGAAGATCCAACTTTTACAATCTATCCTGCAACATACCCTCTATCTGAAACACCAGATGAGCTGAAGAGAACACTTTCAAGTGTTCTTCAAGATGCTGGAAAGGCAAATGTGCAGTTATTCTTTGATGGCACTCTTACAGCTCTTGCCAATGACTTGAGTAGAGGTCTACATGCCAATGGTCACAGATTACTGCTTCAGATGCTAGCTCAAGATAACCCCGAGTTTTGTATTGCATCCCTCCAGAAGTATGGAAGTTTAAGAAACTCGTACCAGAATCGACCCCCTATAGGAATGTCTCTACTGTGGGCATTTGGACAAGGTGGTCTGAAAGACTTCACGGTGGGATTGAAAGTTTGGCAGGAAATCTTTTTACCGGTACTCGAATTGAAGAACTATACGAAATACGTAATAGGATATCTTTCGAAGATAATCGATCATCATTCCGAGATGAATACGATTAATGTTAACCAGGATCAATTCTTTTCTATGATTGACTTGATAAACACAAAGAAGAATGGTTTATCGAAGGAATTTTCTGGTGACCTAATCAAACAATTAAGTAAATATAAGGACATTTATTTCAAGAAGAGCGGCAACAAATTACAAGTTACGTTCAATCAGCTGATGAAGAAACTCCCCAACCAGTACCTAAGCGGTTCGGCCTTGGATCCGTACAACAAAGTGATCATCGAAAGTCTAGTCGACTGTCTGTCTAAAGACGACTCTTGTAACGCCACGTGGCGTCAACTCTTCCACAGGTGCAGTAAGCAGTCTGCCTCACTCCTCGATTTCATAGATTCAAATTGGTTGGAAGTAAGTAAAAAATTGAAATTAAAATCTCTAAAAGCAACGGTGCTACAATATAAAGAAGTGAGCGGAGAAGCTCTTAAGGGCAAGAAAAAGGATGAAGTTCTAGTAAAAACAACGAAGATATGCCAGGATATTCTAGACAGAATGACGAGCACTAGAAGAAATCCCTGGTTGTGGGCCAGCTTTGTTCTGCTGGTCAGCATTGCTGGTCTGGTAGCTTATGACGTGTCTCGGGCTGGAGGAAACTTCCCTAAGAGTACAACGGGAAAGCTGTTCAAAGATTTGGGCATTCTTGAGCATAGTCAACAGGCTTGGCAGAAGACACTGTCCACCTCTGCTCGCGGTTACCTATGGCTTGAAACCAATGCACCGGTTTACTACGCACAAACTGTTGAAATATGTCGTCCATATACACAACTGTCTAAGGACGCATTCTTCATAGCATTAAAGAAAGCTGGTATTCTGTATGGAAATGTCCAAGACTATGTGGTGGAGAAAACGCCTGTCGTTATTAAAACCATTGAAGAGTATGCTCCTGGTCTTGTAGACAACGTTCAGAGCTATGCATCTACGGCCTGGAGCGGACTCAAAAAGTACTCTAGTGACTATTATCAAATCACAACTGACTATTTGGTTACTAAAGTTTTTGTTGGTGATTGGGCCCCAGAGGTACTTCAGAATAAAACGCAGTCGGCTCTGAATATGACAAAGTCCCAAGTTTCATCATATTATGTATGGTTCCGGCAACAGGTTCACATATACTCAGAGATACCTTGA
Protein
MSSGQWEVVGKNKKSQNGKLKSKEDEKKALKNGPKLEDVVPHSQIKSLYAGMDVDGEKKSAKEKKNSDKKGKKQEKKVEPPKPKIPKTIEGALEAIDVTELASIITSNKVRFSNAPLVWLKEVANFLNSKIPIEVEDPTFTIYPATYPLSETPDELKRTLSSVLQDAGKANVQLFFDGTLTALANDLSRGLHANGHRLLLQMLAQDNPEFCIASLQKYGSLRNSYQNRPPIGMSLLWAFGQGGLKDFTVGLKVWQEIFLPVLELKNYTKYVIGYLSKIIDHHSEMNTINVNQDQFFSMIDLINTKKNGLSKEFSGDLIKQLSKYKDIYFKKSGNKLQVTFNQLMKKLPNQYLSGSALDPYNKVIIESLVDCLSKDDSCNATWRQLFHRCSKQSASLLDFIDSNWLEVSKKLKLKSLKATVLQYKEVSGEALKGKKKDEVLVKTTKICQDILDRMTSTRRNPWLWASFVLLVSIAGLVAYDVSRAGGNFPKSTTGKLFKDLGILEHSQQAWQKTLSTSARGYLWLETNAPVYYAQTVEICRPYTQLSKDAFFIALKKAGILYGNVQDYVVEKTPVVIKTIEEYAPGLVDNVQSYASTAWSGLKKYSSDYYQITTDYLVTKVFVGDWAPEVLQNKTQSALNMTKSQVSSYYVWFRQQVHIYSEIP
Summary
Uniprot
H9IX59
A0A2H1VGH5
A0A194QF90
A0A194QVG9
A0A212F095
A0A2A4J9V0
+ More
S4PAR9 A0A0L7KTS8 A0A2J7PI08 A0A067RAK0 A0A1B6C1J5 A0A1Y1NDD2 K7J1Y7 A0A1Q3G1Q5 A0A1B6E3W0 A0A087ZVG6 E2A8B1 A0A2A3ESJ6 E9IHB5 A0A195BSY1 A0A158NNV8 A0A348G6D9 A0A151IX79 F4WVA2 A0A195EX42 A0A310SNZ3 A0A0J7KZ07 A0A0M8ZW21 A0A151WJR9 E2BYY3 A0A1W4XHL7 Q17MR8 A0A1S4EXB5 A0A1B6KGB0 A0A1B6G964 A0A151ICA0 A0A1Q3G1M2 A0A0L7RAA4 A0A1B6IL65 A0A1L8DCB5 A0A1L8DCA9 A0A1B0CKT0 D6WQZ8 A0A1L8DBY2 A0A026WQR4 A0A139WG18 Q7PM66 A0A182KTU8 A0A2C9GPY1 A0A0A1X9C6 A0A034VYR9 A0A0C9RLD5 W8BC69 A0A0M4E0L6 B4MDI3 A0A232EV79 A0A182VJH6 A0A1W4W5N0 A0A0L0CDM6 B3N508 B4HWY3 A0A0J9R010 A0A1I8PUK6 B4P143 A0A3S3NZ24 A0A182TK23 A0A3B0JH49 Q9VKM7 B4KJ59 B4G6Y3 B3MJY8 A0A1I8MN51 Q29CN6 A0A1S3IMQ1 A0A1B0GH77 H9GDR4 A0A3B3DYT5 A0A3P9L7E4 H2MTM5 A0A3P9JH92 A0A3P8RFA8 A0A0F7YZ28 H3B9X2 A0A3N0YM04 M7BBP0 A0A3B3ZIG6 H3B9X1 A0A2U9CUR2 G3NVB0 A0A0B8RVS5 A0A3Q0RYX9 I3JDL0 I3JDL1 A0A3B4H4D6
S4PAR9 A0A0L7KTS8 A0A2J7PI08 A0A067RAK0 A0A1B6C1J5 A0A1Y1NDD2 K7J1Y7 A0A1Q3G1Q5 A0A1B6E3W0 A0A087ZVG6 E2A8B1 A0A2A3ESJ6 E9IHB5 A0A195BSY1 A0A158NNV8 A0A348G6D9 A0A151IX79 F4WVA2 A0A195EX42 A0A310SNZ3 A0A0J7KZ07 A0A0M8ZW21 A0A151WJR9 E2BYY3 A0A1W4XHL7 Q17MR8 A0A1S4EXB5 A0A1B6KGB0 A0A1B6G964 A0A151ICA0 A0A1Q3G1M2 A0A0L7RAA4 A0A1B6IL65 A0A1L8DCB5 A0A1L8DCA9 A0A1B0CKT0 D6WQZ8 A0A1L8DBY2 A0A026WQR4 A0A139WG18 Q7PM66 A0A182KTU8 A0A2C9GPY1 A0A0A1X9C6 A0A034VYR9 A0A0C9RLD5 W8BC69 A0A0M4E0L6 B4MDI3 A0A232EV79 A0A182VJH6 A0A1W4W5N0 A0A0L0CDM6 B3N508 B4HWY3 A0A0J9R010 A0A1I8PUK6 B4P143 A0A3S3NZ24 A0A182TK23 A0A3B0JH49 Q9VKM7 B4KJ59 B4G6Y3 B3MJY8 A0A1I8MN51 Q29CN6 A0A1S3IMQ1 A0A1B0GH77 H9GDR4 A0A3B3DYT5 A0A3P9L7E4 H2MTM5 A0A3P9JH92 A0A3P8RFA8 A0A0F7YZ28 H3B9X2 A0A3N0YM04 M7BBP0 A0A3B3ZIG6 H3B9X1 A0A2U9CUR2 G3NVB0 A0A0B8RVS5 A0A3Q0RYX9 I3JDL0 I3JDL1 A0A3B4H4D6
Pubmed
19121390
26354079
22118469
23622113
26227816
24845553
+ More
28004739 20075255 20798317 21282665 21347285 21719571 17510324 18362917 19820115 24508170 30249741 12364791 20966253 25830018 25348373 24495485 17994087 28648823 26108605 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 15632085 21881562 29451363 17554307 9215903 23624526 25463417 25476704 25186727
28004739 20075255 20798317 21282665 21347285 21719571 17510324 18362917 19820115 24508170 30249741 12364791 20966253 25830018 25348373 24495485 17994087 28648823 26108605 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 15632085 21881562 29451363 17554307 9215903 23624526 25463417 25476704 25186727
EMBL
BABH01018278
ODYU01002171
SOQ39324.1
KQ459053
KPJ04079.1
KQ461073
+ More
KPJ09533.1 AGBW02011140 OWR47153.1 NWSH01002431 PCG68314.1 GAIX01004886 JAA87674.1 JTDY01005773 KOB66647.1 NEVH01025130 PNF15977.1 KK852779 KDR16686.1 GEDC01029922 JAS07376.1 GEZM01009887 JAV94246.1 GFDL01001325 JAV33720.1 GEDC01004686 JAS32612.1 GL437526 EFN70324.1 KZ288193 PBC34186.1 GL763196 EFZ20030.1 KQ976417 KYM89778.1 ADTU01021806 FX986055 BBF98012.1 KQ980843 KYN12297.1 GL888384 EGI61868.1 KQ981953 KYN32449.1 KQ762207 OAD56057.1 LBMM01001920 KMQ95538.1 KQ435845 KOX71196.1 KQ983031 KYQ48123.1 GL451531 EFN79101.1 CH477204 EAT47954.1 GEBQ01029511 JAT10466.1 GECZ01010777 JAS58992.1 KQ978062 KYM97706.1 GFDL01001341 JAV33704.1 KQ414618 KOC67817.1 GECU01020071 JAS87635.1 GFDF01009985 JAV04099.1 GFDF01009986 JAV04098.1 AJWK01016603 KQ971351 EFA06531.2 GFDF01010244 JAV03840.1 KK107128 QOIP01000006 EZA58370.1 RLU21875.1 KYB26829.1 AAAB01008980 EAA14245.5 APCN01001735 GBXI01006423 JAD07869.1 GAKP01011917 JAC47035.1 GBYB01007731 JAG77498.1 GAMC01007710 JAB98845.1 CP012523 ALC38704.1 CH940661 EDW71244.2 NNAY01002039 OXU22256.1 JRES01000536 KNC30351.1 CH954177 EDV58953.1 CH480818 EDW52528.1 CM002910 KMY89652.1 CM000157 EDW88018.1 NCKU01001492 RWS11991.1 OUUW01000006 SPP81724.1 AE014134 BT003656 AAF53037.2 AAO39660.1 CH933807 EDW11421.1 CH479180 EDW28302.1 CH902620 EDV32443.1 CH475442 EAL29396.2 AJWK01004026 AJWK01004027 AJWK01004028 AJWK01004029 AAWZ02030759 GBEW01001090 JAI09275.1 AFYH01030204 AFYH01030205 AFYH01030206 AFYH01030207 AFYH01030208 AFYH01030209 RJVU01035944 ROL47233.1 KB537686 EMP32980.1 CP026262 AWP20331.1 GBSH01002850 JAG66177.1 AERX01015145
KPJ09533.1 AGBW02011140 OWR47153.1 NWSH01002431 PCG68314.1 GAIX01004886 JAA87674.1 JTDY01005773 KOB66647.1 NEVH01025130 PNF15977.1 KK852779 KDR16686.1 GEDC01029922 JAS07376.1 GEZM01009887 JAV94246.1 GFDL01001325 JAV33720.1 GEDC01004686 JAS32612.1 GL437526 EFN70324.1 KZ288193 PBC34186.1 GL763196 EFZ20030.1 KQ976417 KYM89778.1 ADTU01021806 FX986055 BBF98012.1 KQ980843 KYN12297.1 GL888384 EGI61868.1 KQ981953 KYN32449.1 KQ762207 OAD56057.1 LBMM01001920 KMQ95538.1 KQ435845 KOX71196.1 KQ983031 KYQ48123.1 GL451531 EFN79101.1 CH477204 EAT47954.1 GEBQ01029511 JAT10466.1 GECZ01010777 JAS58992.1 KQ978062 KYM97706.1 GFDL01001341 JAV33704.1 KQ414618 KOC67817.1 GECU01020071 JAS87635.1 GFDF01009985 JAV04099.1 GFDF01009986 JAV04098.1 AJWK01016603 KQ971351 EFA06531.2 GFDF01010244 JAV03840.1 KK107128 QOIP01000006 EZA58370.1 RLU21875.1 KYB26829.1 AAAB01008980 EAA14245.5 APCN01001735 GBXI01006423 JAD07869.1 GAKP01011917 JAC47035.1 GBYB01007731 JAG77498.1 GAMC01007710 JAB98845.1 CP012523 ALC38704.1 CH940661 EDW71244.2 NNAY01002039 OXU22256.1 JRES01000536 KNC30351.1 CH954177 EDV58953.1 CH480818 EDW52528.1 CM002910 KMY89652.1 CM000157 EDW88018.1 NCKU01001492 RWS11991.1 OUUW01000006 SPP81724.1 AE014134 BT003656 AAF53037.2 AAO39660.1 CH933807 EDW11421.1 CH479180 EDW28302.1 CH902620 EDV32443.1 CH475442 EAL29396.2 AJWK01004026 AJWK01004027 AJWK01004028 AJWK01004029 AAWZ02030759 GBEW01001090 JAI09275.1 AFYH01030204 AFYH01030205 AFYH01030206 AFYH01030207 AFYH01030208 AFYH01030209 RJVU01035944 ROL47233.1 KB537686 EMP32980.1 CP026262 AWP20331.1 GBSH01002850 JAG66177.1 AERX01015145
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000218220
UP000037510
+ More
UP000235965 UP000027135 UP000002358 UP000005203 UP000000311 UP000242457 UP000078540 UP000005205 UP000078492 UP000007755 UP000078541 UP000036403 UP000053105 UP000075809 UP000008237 UP000192223 UP000008820 UP000078542 UP000053825 UP000092461 UP000007266 UP000053097 UP000279307 UP000007062 UP000075882 UP000075840 UP000092553 UP000008792 UP000215335 UP000075903 UP000192221 UP000037069 UP000008711 UP000001292 UP000095300 UP000002282 UP000285301 UP000075902 UP000268350 UP000000803 UP000009192 UP000008744 UP000007801 UP000095301 UP000001819 UP000085678 UP000001646 UP000261560 UP000265180 UP000001038 UP000265200 UP000265100 UP000008672 UP000031443 UP000261520 UP000246464 UP000007635 UP000261340 UP000005207 UP000261460
UP000235965 UP000027135 UP000002358 UP000005203 UP000000311 UP000242457 UP000078540 UP000005205 UP000078492 UP000007755 UP000078541 UP000036403 UP000053105 UP000075809 UP000008237 UP000192223 UP000008820 UP000078542 UP000053825 UP000092461 UP000007266 UP000053097 UP000279307 UP000007062 UP000075882 UP000075840 UP000092553 UP000008792 UP000215335 UP000075903 UP000192221 UP000037069 UP000008711 UP000001292 UP000095300 UP000002282 UP000285301 UP000075902 UP000268350 UP000000803 UP000009192 UP000008744 UP000007801 UP000095301 UP000001819 UP000085678 UP000001646 UP000261560 UP000265180 UP000001038 UP000265200 UP000265100 UP000008672 UP000031443 UP000261520 UP000246464 UP000007635 UP000261340 UP000005207 UP000261460
Pfam
PF10151 TMEM214
SUPFAM
SSF48371
SSF48371
ProteinModelPortal
H9IX59
A0A2H1VGH5
A0A194QF90
A0A194QVG9
A0A212F095
A0A2A4J9V0
+ More
S4PAR9 A0A0L7KTS8 A0A2J7PI08 A0A067RAK0 A0A1B6C1J5 A0A1Y1NDD2 K7J1Y7 A0A1Q3G1Q5 A0A1B6E3W0 A0A087ZVG6 E2A8B1 A0A2A3ESJ6 E9IHB5 A0A195BSY1 A0A158NNV8 A0A348G6D9 A0A151IX79 F4WVA2 A0A195EX42 A0A310SNZ3 A0A0J7KZ07 A0A0M8ZW21 A0A151WJR9 E2BYY3 A0A1W4XHL7 Q17MR8 A0A1S4EXB5 A0A1B6KGB0 A0A1B6G964 A0A151ICA0 A0A1Q3G1M2 A0A0L7RAA4 A0A1B6IL65 A0A1L8DCB5 A0A1L8DCA9 A0A1B0CKT0 D6WQZ8 A0A1L8DBY2 A0A026WQR4 A0A139WG18 Q7PM66 A0A182KTU8 A0A2C9GPY1 A0A0A1X9C6 A0A034VYR9 A0A0C9RLD5 W8BC69 A0A0M4E0L6 B4MDI3 A0A232EV79 A0A182VJH6 A0A1W4W5N0 A0A0L0CDM6 B3N508 B4HWY3 A0A0J9R010 A0A1I8PUK6 B4P143 A0A3S3NZ24 A0A182TK23 A0A3B0JH49 Q9VKM7 B4KJ59 B4G6Y3 B3MJY8 A0A1I8MN51 Q29CN6 A0A1S3IMQ1 A0A1B0GH77 H9GDR4 A0A3B3DYT5 A0A3P9L7E4 H2MTM5 A0A3P9JH92 A0A3P8RFA8 A0A0F7YZ28 H3B9X2 A0A3N0YM04 M7BBP0 A0A3B3ZIG6 H3B9X1 A0A2U9CUR2 G3NVB0 A0A0B8RVS5 A0A3Q0RYX9 I3JDL0 I3JDL1 A0A3B4H4D6
S4PAR9 A0A0L7KTS8 A0A2J7PI08 A0A067RAK0 A0A1B6C1J5 A0A1Y1NDD2 K7J1Y7 A0A1Q3G1Q5 A0A1B6E3W0 A0A087ZVG6 E2A8B1 A0A2A3ESJ6 E9IHB5 A0A195BSY1 A0A158NNV8 A0A348G6D9 A0A151IX79 F4WVA2 A0A195EX42 A0A310SNZ3 A0A0J7KZ07 A0A0M8ZW21 A0A151WJR9 E2BYY3 A0A1W4XHL7 Q17MR8 A0A1S4EXB5 A0A1B6KGB0 A0A1B6G964 A0A151ICA0 A0A1Q3G1M2 A0A0L7RAA4 A0A1B6IL65 A0A1L8DCB5 A0A1L8DCA9 A0A1B0CKT0 D6WQZ8 A0A1L8DBY2 A0A026WQR4 A0A139WG18 Q7PM66 A0A182KTU8 A0A2C9GPY1 A0A0A1X9C6 A0A034VYR9 A0A0C9RLD5 W8BC69 A0A0M4E0L6 B4MDI3 A0A232EV79 A0A182VJH6 A0A1W4W5N0 A0A0L0CDM6 B3N508 B4HWY3 A0A0J9R010 A0A1I8PUK6 B4P143 A0A3S3NZ24 A0A182TK23 A0A3B0JH49 Q9VKM7 B4KJ59 B4G6Y3 B3MJY8 A0A1I8MN51 Q29CN6 A0A1S3IMQ1 A0A1B0GH77 H9GDR4 A0A3B3DYT5 A0A3P9L7E4 H2MTM5 A0A3P9JH92 A0A3P8RFA8 A0A0F7YZ28 H3B9X2 A0A3N0YM04 M7BBP0 A0A3B3ZIG6 H3B9X1 A0A2U9CUR2 G3NVB0 A0A0B8RVS5 A0A3Q0RYX9 I3JDL0 I3JDL1 A0A3B4H4D6
Ontologies
PANTHER
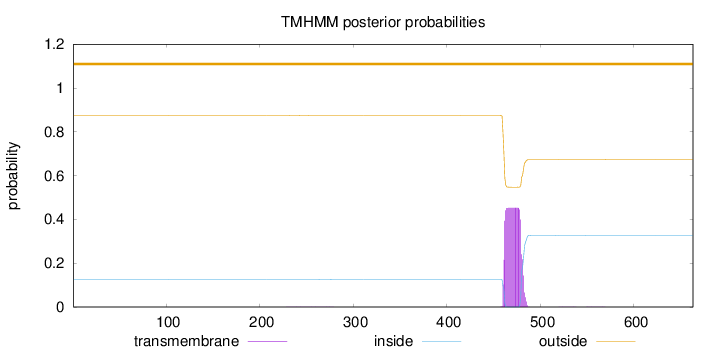
Topology
Length:
663
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
9.11726999999998
Exp number, first 60 AAs:
0
Total prob of N-in:
0.12427
outside
1 - 663
Population Genetic Test Statistics
Pi
254.110028
Theta
185.456829
Tajima's D
1.157303
CLR
0.289748
CSRT
0.699765011749413
Interpretation
Uncertain
