Gene
KWMTBOMO11313 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002071
Annotation
PREDICTED:_5-formyltetrahydrofolate_cyclo-ligase-like_isoform_X2_[Amyelois_transitella]
Full name
5-formyltetrahydrofolate cyclo-ligase
Alternative Name
5,10-methenyl-tetrahydrofolate synthetase
Location in the cell
Mitochondrial Reliability : 2.143
Sequence
CDS
ATGAGTCCAATAAAACCAAATCCTGCCAAAGCCTTGCTACGTAACGAAATTGCCGCTAAAATCGCCGCCTTGACGAATGAAGAGAAAAAAAGACAATCTCAAATTGTTTATGAAAAGGTTATCAACCACTCATGGTACAAATCATCAAGTCGTATTGCTCTCTACATGAGCACCGAAAATGAGATAGACACTGCTCCTCTAATCAAACACATCCAAGCCAGAGGGGCTGCGGCCTTCGTGCCACAGTACGCCGGAGGGCGCATGAGGATGTTACACTTGGAGACCGGTGATGAGCAGACCATGCCAAAAACTAAACACGGGATATCTCAGCATGGGAAAGACCAAGCCAGGGACGATGCTCTAGAAACAGGTGGATTAGATCTGATCATAGCTCCAGGGGTGGCGTTTTCTCGTTCCGGAGACAGGCTGGGACACGGAGGTGGATACTACGACAAGTTCATCACTAATTTGAGGCTCAATCCAGAAACAGCTCCGAAGATAGTTGCAGTGGCTTTTAACTGTCAAGTTGTCGACGAGGTGCCGACTAACGAACAAGACCAGAAAGTGGACGAGGTTATATTCGCTGAATAA
Protein
MSPIKPNPAKALLRNEIAAKIAALTNEEKKRQSQIVYEKVINHSWYKSSSRIALYMSTENEIDTAPLIKHIQARGAAAFVPQYAGGRMRMLHLETGDEQTMPKTKHGISQHGKDQARDDALETGGLDLIIAPGVAFSRSGDRLGHGGGYYDKFITNLRLNPETAPKIVAVAFNCQVVDEVPTNEQDQKVDEVIFAE
Summary
Description
Contributes to tetrahydrofolate metabolism. Helps regulate carbon flow through the folate-dependent one-carbon metabolic network that supplies carbon for the biosynthesis of purines, thymidine and amino acids. Catalyzes the irreversible conversion of 5-formyltetrahydrofolate (5-FTHF) to yield 5,10-methenyltetrahydrofolate.
Contributes to tetrahydrofolate metabolism. Helps regulate carbon flow through the folate-dependent one-carbon metabolic network that supplies carbon for the biosynthesis of purines, thymidine and amino acids. Catalyzes the irreversible conversion of 5-formyltetrahydrofolate (5-CHO-H(4)PteGlu) to yield 5,10-methenyltetrahydrofolate (By similarity).
Contributes to tetrahydrofolate metabolism. Helps regulate carbon flow through the folate-dependent one-carbon metabolic network that supplies carbon for the biosynthesis of purines, thymidine and amino acids. Catalyzes the irreversible conversion of 5-formyltetrahydrofolate (5-CHO-H(4)PteGlu) to yield 5,10-methenyltetrahydrofolate (By similarity).
Catalytic Activity
5-formyltetrahydrofolate + ATP = 5,10-methenyltetrahydrofolate + ADP + phosphate
Cofactor
Mg(2+)
Subunit
Monomer.
Similarity
Belongs to the 5-formyltetrahydrofolate cyclo-ligase family.
Keywords
3D-structure
Acetylation
Alternative splicing
ATP-binding
Complete proteome
Cytoplasm
Folate-binding
Ligase
Magnesium
Metal-binding
Nucleotide-binding
Polymorphism
Reference proteome
Feature
chain 5-formyltetrahydrofolate cyclo-ligase
splice variant In isoform 2.
sequence variant In dbSNP:rs8923.
splice variant In isoform 2.
sequence variant In dbSNP:rs8923.
Uniprot
H9IXT8
A0A2H1WK73
A0A2W1BHP1
A0A2A4IWE7
A0A2A4IY02
A0A3S2NXC3
+ More
A0A194QGL2 A0A194QVN7 A0A212FFP3 J3JVY5 A0A232FEM7 K7JH06 U4U6T3 D6WQI7 A0A0C9RNG4 A0A1W4X470 A0A3L8D8P2 N6THY6 A0A2G8JXW3 A0A088A7I3 A0A0M8ZMZ8 A0A151IJY0 A0A2A3ED96 A0A026X241 A0A0D9RRA7 A0A0P4WHX0 A0A2U3Y8J2 A0A151WTA5 A0A195EHD9 A0A2Y9GE88 A0A2K5SGM8 A0A2K5LTS3 G7P997 F6VVW7 F6YZ79 A0A096NMX4 A0A1S3JK03 U3CWB8 H2NNZ8 A0A0P4WD20 U3FH71 A0A2K5D223 A0A2K6MAJ0 A0A2K6PB55 F6SFK7 A0A2I3FPQ7 A0A2I2UGT6 A0A0J7KQF3 A0A195B048 P49914 G3R914 A0A2K6A0D3 A0A2K5JX60 H2Q9X5 A0A158NKF9 U5EYE0 A0A2T7NFU6 A0A2U3W637 A0A1B6CAN7 A0A2K6CEG9 A0A1B6DQ05 Q96EE9 D2I5Q7 A0A0L7QNH6 A0A3Q7QT93 A0A1U7TBM6 G1LCH4 G3GTB3 I3MCZ0 F7C3A9 A0A3B5KFU0 A0A0N8EUE8 G3T1C1 A0A1A6HUR1 L7N466 A0A154PMF8 A0A1Y1MCK7 Q9D110 A0A3B4DW17 E2RR20 A0A3Q7WX09 H3ABJ5 Q3TYX5 A0A250YBY9 A0A3Q3M0N9 A0A3Q7SHP9 Q5M9F6 A0A0G2L0J2 A0A226DK10 A0A2Y9LNF9 A0A3P8V7K9 A0A341BA42 A0A2U4C7R9 A0A340WJ40 A0A384B3Z1 R7T3G1 A0A210PY18 A0A3B5AGJ6 A0A3B1JLE9 H0X584
A0A194QGL2 A0A194QVN7 A0A212FFP3 J3JVY5 A0A232FEM7 K7JH06 U4U6T3 D6WQI7 A0A0C9RNG4 A0A1W4X470 A0A3L8D8P2 N6THY6 A0A2G8JXW3 A0A088A7I3 A0A0M8ZMZ8 A0A151IJY0 A0A2A3ED96 A0A026X241 A0A0D9RRA7 A0A0P4WHX0 A0A2U3Y8J2 A0A151WTA5 A0A195EHD9 A0A2Y9GE88 A0A2K5SGM8 A0A2K5LTS3 G7P997 F6VVW7 F6YZ79 A0A096NMX4 A0A1S3JK03 U3CWB8 H2NNZ8 A0A0P4WD20 U3FH71 A0A2K5D223 A0A2K6MAJ0 A0A2K6PB55 F6SFK7 A0A2I3FPQ7 A0A2I2UGT6 A0A0J7KQF3 A0A195B048 P49914 G3R914 A0A2K6A0D3 A0A2K5JX60 H2Q9X5 A0A158NKF9 U5EYE0 A0A2T7NFU6 A0A2U3W637 A0A1B6CAN7 A0A2K6CEG9 A0A1B6DQ05 Q96EE9 D2I5Q7 A0A0L7QNH6 A0A3Q7QT93 A0A1U7TBM6 G1LCH4 G3GTB3 I3MCZ0 F7C3A9 A0A3B5KFU0 A0A0N8EUE8 G3T1C1 A0A1A6HUR1 L7N466 A0A154PMF8 A0A1Y1MCK7 Q9D110 A0A3B4DW17 E2RR20 A0A3Q7WX09 H3ABJ5 Q3TYX5 A0A250YBY9 A0A3Q3M0N9 A0A3Q7SHP9 Q5M9F6 A0A0G2L0J2 A0A226DK10 A0A2Y9LNF9 A0A3P8V7K9 A0A341BA42 A0A2U4C7R9 A0A340WJ40 A0A384B3Z1 R7T3G1 A0A210PY18 A0A3B5AGJ6 A0A3B1JLE9 H0X584
EC Number
6.3.3.2
Pubmed
19121390
28756777
26354079
22118469
22516182
28648823
+ More
20075255 23537049 18362917 19820115 30249741 29023486 24508170 22002653 17431167 25319552 12481130 15114417 25243066 25362486 17495919 17975172 8522195 16572171 15489334 3801490 19413330 22814378 19738041 16136131 21347285 20010809 21804562 19892987 21551351 19468303 21183079 28004739 16141072 16341006 9215903 10349636 11042159 11076861 11217851 12466851 16141073 28087693 15057822 15632090 23594743 24487278 23254933 28812685 25329095
20075255 23537049 18362917 19820115 30249741 29023486 24508170 22002653 17431167 25319552 12481130 15114417 25243066 25362486 17495919 17975172 8522195 16572171 15489334 3801490 19413330 22814378 19738041 16136131 21347285 20010809 21804562 19892987 21551351 19468303 21183079 28004739 16141072 16341006 9215903 10349636 11042159 11076861 11217851 12466851 16141073 28087693 15057822 15632090 23594743 24487278 23254933 28812685 25329095
EMBL
BABH01015585
BABH01015586
BABH01015587
ODYU01009166
SOQ53407.1
KZ150054
+ More
PZC74358.1 NWSH01005325 PCG64287.1 PCG64288.1 RSAL01000034 RVE51390.1 KQ459053 KPJ04075.1 KQ461073 KPJ09537.1 AGBW02008791 OWR52554.1 BT127403 AEE62365.1 NNAY01000332 OXU29145.1 KB632095 ERL88777.1 KQ971354 EFA06068.1 GBYB01008621 JAG78388.1 QOIP01000011 RLU16674.1 APGK01025866 KB740605 ENN80019.1 MRZV01001107 PIK40573.1 KQ436033 KOX67535.1 KQ977294 KYN03946.1 KZ288279 PBC29688.1 KK107021 EZA62385.1 AQIB01145754 GDRN01049135 JAI66603.1 KQ982762 KYQ51063.1 KQ978881 KYN27688.1 AQIA01063441 AQIA01063442 AQIA01063443 CM001282 EHH63272.1 JSUE03038848 JSUE03038849 JSUE03038850 JU323084 JU474017 JV045948 CM001259 AFE66840.1 AFH30821.1 AFI36019.1 EHH27528.1 AHZZ02031365 AHZZ02031366 GAMT01007704 GAMS01009331 GAMR01010065 GAMP01007133 JAB04157.1 JAB13805.1 JAB23867.1 JAB45622.1 ABGA01219171 ABGA01219172 ABGA01269314 ABGA01269315 ABGA01269316 ABGA01269317 ABGA01269318 ABGA01269319 GDRN01049134 JAI66604.1 GAMQ01004356 JAB37495.1 ADFV01087605 ADFV01087606 ADFV01087607 AANG04002993 LBMM01004329 KMQ92563.1 KQ976692 KYM77831.1 L38928 AC015871 AC021483 BC019921 CK002935 CABD030097388 CABD030097389 CABD030097390 AACZ04052725 GABC01008601 GABF01010635 GABD01005894 GABE01009334 JAA02737.1 JAA11510.1 JAA27206.1 JAA35405.1 ADTU01018821 GANO01002057 JAB57814.1 PZQS01000013 PVD20002.1 GEDC01026750 GEDC01018168 GEDC01014074 GEDC01004956 GEDC01003269 JAS10548.1 JAS19130.1 JAS23224.1 JAS32342.1 JAS34029.1 GEDC01031681 GEDC01029328 GEDC01027988 GEDC01027181 GEDC01017528 GEDC01011532 GEDC01009548 GEDC01007612 JAS05617.1 JAS07970.1 JAS09310.1 JAS10117.1 JAS19770.1 JAS25766.1 JAS27750.1 JAS29686.1 BC012417 AAH12417.1 GL194762 EFB25996.1 KQ414851 KOC60180.1 ACTA01180869 ACTA01188867 ACTA01196865 JH000018 EGV94500.1 AGTP01003694 AGTP01003695 AGTP01003696 GEBF01001779 JAO01854.1 LZPO01008151 OBS82188.1 AC163666 CT030710 KQ434978 KZC13023.1 GEZM01035129 GEZM01035128 JAV83393.1 AK004092 AC135634 CT030686 AAEX03002384 AFYH01020396 AFYH01020397 AFYH01020398 AFYH01020399 AK158264 BAE34435.1 GFFW01003651 JAV41137.1 AC120938 BC087144 CH473954 AAH87144.1 EDL77576.1 CABZ01029544 CABZ01029545 CABZ01029546 CU695017 LNIX01000017 OXA45460.1 AMQN01003670 KB312450 ELT87198.1 NEDP02005404 OWF41377.1 AAQR03026739 AAQR03026740 AAQR03026741
PZC74358.1 NWSH01005325 PCG64287.1 PCG64288.1 RSAL01000034 RVE51390.1 KQ459053 KPJ04075.1 KQ461073 KPJ09537.1 AGBW02008791 OWR52554.1 BT127403 AEE62365.1 NNAY01000332 OXU29145.1 KB632095 ERL88777.1 KQ971354 EFA06068.1 GBYB01008621 JAG78388.1 QOIP01000011 RLU16674.1 APGK01025866 KB740605 ENN80019.1 MRZV01001107 PIK40573.1 KQ436033 KOX67535.1 KQ977294 KYN03946.1 KZ288279 PBC29688.1 KK107021 EZA62385.1 AQIB01145754 GDRN01049135 JAI66603.1 KQ982762 KYQ51063.1 KQ978881 KYN27688.1 AQIA01063441 AQIA01063442 AQIA01063443 CM001282 EHH63272.1 JSUE03038848 JSUE03038849 JSUE03038850 JU323084 JU474017 JV045948 CM001259 AFE66840.1 AFH30821.1 AFI36019.1 EHH27528.1 AHZZ02031365 AHZZ02031366 GAMT01007704 GAMS01009331 GAMR01010065 GAMP01007133 JAB04157.1 JAB13805.1 JAB23867.1 JAB45622.1 ABGA01219171 ABGA01219172 ABGA01269314 ABGA01269315 ABGA01269316 ABGA01269317 ABGA01269318 ABGA01269319 GDRN01049134 JAI66604.1 GAMQ01004356 JAB37495.1 ADFV01087605 ADFV01087606 ADFV01087607 AANG04002993 LBMM01004329 KMQ92563.1 KQ976692 KYM77831.1 L38928 AC015871 AC021483 BC019921 CK002935 CABD030097388 CABD030097389 CABD030097390 AACZ04052725 GABC01008601 GABF01010635 GABD01005894 GABE01009334 JAA02737.1 JAA11510.1 JAA27206.1 JAA35405.1 ADTU01018821 GANO01002057 JAB57814.1 PZQS01000013 PVD20002.1 GEDC01026750 GEDC01018168 GEDC01014074 GEDC01004956 GEDC01003269 JAS10548.1 JAS19130.1 JAS23224.1 JAS32342.1 JAS34029.1 GEDC01031681 GEDC01029328 GEDC01027988 GEDC01027181 GEDC01017528 GEDC01011532 GEDC01009548 GEDC01007612 JAS05617.1 JAS07970.1 JAS09310.1 JAS10117.1 JAS19770.1 JAS25766.1 JAS27750.1 JAS29686.1 BC012417 AAH12417.1 GL194762 EFB25996.1 KQ414851 KOC60180.1 ACTA01180869 ACTA01188867 ACTA01196865 JH000018 EGV94500.1 AGTP01003694 AGTP01003695 AGTP01003696 GEBF01001779 JAO01854.1 LZPO01008151 OBS82188.1 AC163666 CT030710 KQ434978 KZC13023.1 GEZM01035129 GEZM01035128 JAV83393.1 AK004092 AC135634 CT030686 AAEX03002384 AFYH01020396 AFYH01020397 AFYH01020398 AFYH01020399 AK158264 BAE34435.1 GFFW01003651 JAV41137.1 AC120938 BC087144 CH473954 AAH87144.1 EDL77576.1 CABZ01029544 CABZ01029545 CABZ01029546 CU695017 LNIX01000017 OXA45460.1 AMQN01003670 KB312450 ELT87198.1 NEDP02005404 OWF41377.1 AAQR03026739 AAQR03026740 AAQR03026741
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000007151
+ More
UP000215335 UP000002358 UP000030742 UP000007266 UP000192223 UP000279307 UP000019118 UP000230750 UP000005203 UP000053105 UP000078542 UP000242457 UP000053097 UP000029965 UP000245341 UP000075809 UP000078492 UP000248481 UP000233040 UP000233060 UP000009130 UP000233100 UP000006718 UP000008144 UP000028761 UP000085678 UP000008225 UP000001595 UP000233020 UP000233180 UP000233200 UP000002280 UP000001073 UP000011712 UP000036403 UP000078540 UP000005640 UP000001519 UP000233140 UP000233080 UP000002277 UP000005205 UP000245119 UP000245340 UP000233120 UP000053825 UP000286641 UP000189704 UP000008912 UP000001075 UP000005215 UP000002281 UP000005226 UP000007646 UP000092124 UP000000589 UP000076502 UP000261440 UP000002254 UP000286642 UP000008672 UP000261640 UP000286640 UP000002494 UP000000437 UP000198287 UP000248483 UP000265120 UP000252040 UP000245320 UP000265300 UP000261681 UP000014760 UP000242188 UP000261400 UP000018467 UP000005225
UP000215335 UP000002358 UP000030742 UP000007266 UP000192223 UP000279307 UP000019118 UP000230750 UP000005203 UP000053105 UP000078542 UP000242457 UP000053097 UP000029965 UP000245341 UP000075809 UP000078492 UP000248481 UP000233040 UP000233060 UP000009130 UP000233100 UP000006718 UP000008144 UP000028761 UP000085678 UP000008225 UP000001595 UP000233020 UP000233180 UP000233200 UP000002280 UP000001073 UP000011712 UP000036403 UP000078540 UP000005640 UP000001519 UP000233140 UP000233080 UP000002277 UP000005205 UP000245119 UP000245340 UP000233120 UP000053825 UP000286641 UP000189704 UP000008912 UP000001075 UP000005215 UP000002281 UP000005226 UP000007646 UP000092124 UP000000589 UP000076502 UP000261440 UP000002254 UP000286642 UP000008672 UP000261640 UP000286640 UP000002494 UP000000437 UP000198287 UP000248483 UP000265120 UP000252040 UP000245320 UP000265300 UP000261681 UP000014760 UP000242188 UP000261400 UP000018467 UP000005225
Pfam
PF01812 5-FTHF_cyc-lig
Interpro
SUPFAM
SSF100950
SSF100950
Gene 3D
ProteinModelPortal
H9IXT8
A0A2H1WK73
A0A2W1BHP1
A0A2A4IWE7
A0A2A4IY02
A0A3S2NXC3
+ More
A0A194QGL2 A0A194QVN7 A0A212FFP3 J3JVY5 A0A232FEM7 K7JH06 U4U6T3 D6WQI7 A0A0C9RNG4 A0A1W4X470 A0A3L8D8P2 N6THY6 A0A2G8JXW3 A0A088A7I3 A0A0M8ZMZ8 A0A151IJY0 A0A2A3ED96 A0A026X241 A0A0D9RRA7 A0A0P4WHX0 A0A2U3Y8J2 A0A151WTA5 A0A195EHD9 A0A2Y9GE88 A0A2K5SGM8 A0A2K5LTS3 G7P997 F6VVW7 F6YZ79 A0A096NMX4 A0A1S3JK03 U3CWB8 H2NNZ8 A0A0P4WD20 U3FH71 A0A2K5D223 A0A2K6MAJ0 A0A2K6PB55 F6SFK7 A0A2I3FPQ7 A0A2I2UGT6 A0A0J7KQF3 A0A195B048 P49914 G3R914 A0A2K6A0D3 A0A2K5JX60 H2Q9X5 A0A158NKF9 U5EYE0 A0A2T7NFU6 A0A2U3W637 A0A1B6CAN7 A0A2K6CEG9 A0A1B6DQ05 Q96EE9 D2I5Q7 A0A0L7QNH6 A0A3Q7QT93 A0A1U7TBM6 G1LCH4 G3GTB3 I3MCZ0 F7C3A9 A0A3B5KFU0 A0A0N8EUE8 G3T1C1 A0A1A6HUR1 L7N466 A0A154PMF8 A0A1Y1MCK7 Q9D110 A0A3B4DW17 E2RR20 A0A3Q7WX09 H3ABJ5 Q3TYX5 A0A250YBY9 A0A3Q3M0N9 A0A3Q7SHP9 Q5M9F6 A0A0G2L0J2 A0A226DK10 A0A2Y9LNF9 A0A3P8V7K9 A0A341BA42 A0A2U4C7R9 A0A340WJ40 A0A384B3Z1 R7T3G1 A0A210PY18 A0A3B5AGJ6 A0A3B1JLE9 H0X584
A0A194QGL2 A0A194QVN7 A0A212FFP3 J3JVY5 A0A232FEM7 K7JH06 U4U6T3 D6WQI7 A0A0C9RNG4 A0A1W4X470 A0A3L8D8P2 N6THY6 A0A2G8JXW3 A0A088A7I3 A0A0M8ZMZ8 A0A151IJY0 A0A2A3ED96 A0A026X241 A0A0D9RRA7 A0A0P4WHX0 A0A2U3Y8J2 A0A151WTA5 A0A195EHD9 A0A2Y9GE88 A0A2K5SGM8 A0A2K5LTS3 G7P997 F6VVW7 F6YZ79 A0A096NMX4 A0A1S3JK03 U3CWB8 H2NNZ8 A0A0P4WD20 U3FH71 A0A2K5D223 A0A2K6MAJ0 A0A2K6PB55 F6SFK7 A0A2I3FPQ7 A0A2I2UGT6 A0A0J7KQF3 A0A195B048 P49914 G3R914 A0A2K6A0D3 A0A2K5JX60 H2Q9X5 A0A158NKF9 U5EYE0 A0A2T7NFU6 A0A2U3W637 A0A1B6CAN7 A0A2K6CEG9 A0A1B6DQ05 Q96EE9 D2I5Q7 A0A0L7QNH6 A0A3Q7QT93 A0A1U7TBM6 G1LCH4 G3GTB3 I3MCZ0 F7C3A9 A0A3B5KFU0 A0A0N8EUE8 G3T1C1 A0A1A6HUR1 L7N466 A0A154PMF8 A0A1Y1MCK7 Q9D110 A0A3B4DW17 E2RR20 A0A3Q7WX09 H3ABJ5 Q3TYX5 A0A250YBY9 A0A3Q3M0N9 A0A3Q7SHP9 Q5M9F6 A0A0G2L0J2 A0A226DK10 A0A2Y9LNF9 A0A3P8V7K9 A0A341BA42 A0A2U4C7R9 A0A340WJ40 A0A384B3Z1 R7T3G1 A0A210PY18 A0A3B5AGJ6 A0A3B1JLE9 H0X584
PDB
3HY6
E-value=1.44929e-38,
Score=396
Ontologies
PATHWAY
GO
PANTHER
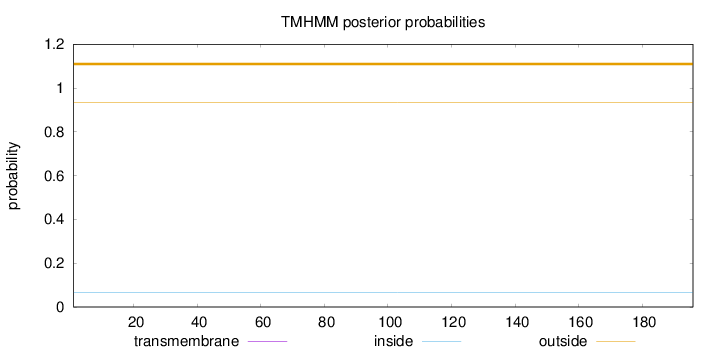
Topology
Subcellular location
Cytoplasm
Length:
196
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00167
Exp number, first 60 AAs:
0
Total prob of N-in:
0.06537
outside
1 - 196
Population Genetic Test Statistics
Pi
247.549598
Theta
170.499114
Tajima's D
1.297799
CLR
0.387179
CSRT
0.743712814359282
Interpretation
Uncertain
