Gene
KWMTBOMO11311 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001841
Annotation
Carboxylesterase_ae27_[Operophtera_brumata]
Full name
Carboxylic ester hydrolase
Location in the cell
PlasmaMembrane Reliability : 3.519
Sequence
CDS
ATGTTTGTTTTACTAATCGCGTTCGTGTCGTTCACGTCCGCCCAAGATAGTCTTCAAGTGAACACCACTGAGGGTACGGTGGAAGGCAGCAGAGCGGCTGACGGAGATTATTTGACTTTCTACGATATACCATACGCCGGTCCAACATCTGGAGAGAACAGATTCAAGGCTCCTAGTCCGCCAACGAACTACTCTGGAGTGTATCATGCCGTCAATCGGAATATTTTGTGCGCCCAGCCCAACACCCGGGGCCTTATTGGAGTAGAGAACTGTCTGACACTCAGCATATACACGAAAAATACGACCACACCCAAACCGGTTTTCGTTTGGCTCAACGCTGAACAATACGCCTCCACCACCACACCAGTGTTCAGCTACAAGAAAATCGTCGAAGAGAACATAGTTTTCGTGCCTCTGAACTTCCGTCTATCAATCTTCGGTTTCATTTGCCTCGGTGTTCCCGACGCACCAGGCAACGCTGGTCTCAAAGACATCCTCCAAGGTCTGACCTGGCTGAAAAGCAACATCGCTGGATTCGGCGGGGACCCTAACAATATTGTGCTTATTGGTCATGGTTCGGGCGCAGCGCTAGTAGATCTGTTGACCATGTCTCCGAGATCCAAGAACCTCGTCCACAAAGCTATTGCACTGAGTGGATCTGCTCTATCACCGTGGGCAGTAGCATATGAGCCTGTACGCTACGCCGAAGTATTTGGGGGGGTGTTGGGAATCACACAGAAATCTAGAGAGCAATTAGCGAAACTTCTTCAGACTACGAATCCAAACGTACTTGCTACAGCGCTAGAAGAATTCAAATTCACAAACAACAGTCTCATATTCGCGCCTTGCATCGAGAGTCAAAATAGTGATCCTAATGAGACAATCATAACAGATGCGCCATTGAACATTTTGCGTTCCGGGAATTACAATCACGTTCCTTACATCGCCGGTTTCACGACTAGAGAGGGCACGTTGAGGGCTTCCGAAGTTGTCTTTAATAGATGGCTCGAGGGTATGGAAGCGAATTTCACAAATTTCCTTCCAGTGGATTTAGATACGAACGGAAACGCAACTGCCGTTCAAGATATAAAGAGATTTTACTTTGGCGATAGAACGATTGACATGGGAACCATCGAGGACTATTTAGAGTACCAGGGCGATACACTTATTTTATTGGCCGTGATAAGAACGGCTAGGGAACGGGGAGTAACTTCCACGAGTCCTGTGAGACTATTAGAATTCGGCTACAGAGGAACACTGAACTCCGATTGGCCCTTCAACCAGATCCCCCTGCACGGAGTGAAGCACGGTGGATTTCTGAACTACCTCTTCGACTATGACTTGCGACCTTCAGACGTTCAGGCGATGAACTCTCTTGTGAGACGTCTAACAAGATTTGCACACACCGGAGAGCCAGCAAACTCGGCCGCTGCAAATGTGGTTTGGTCCGCAGTGGACAAAGCCAATTTGAATTACCTCTACTACGGAGGAAACGATGGGACTGGCTCTGTGGCCTACGTCGAAGAAGACAGGGTCAACCCGCACACGACTAGGATGACATTCTGGAATTCGACCTACGAGAGATATTACGTTCCGCCAAGACCTGTCTCCGGGGCCAACAGCCTTCTCGGCGTAGCTCTGTTTGTTACGCTAAGTCAACTGTTCTTCTTAATGTTTTAG
Protein
MFVLLIAFVSFTSAQDSLQVNTTEGTVEGSRAADGDYLTFYDIPYAGPTSGENRFKAPSPPTNYSGVYHAVNRNILCAQPNTRGLIGVENCLTLSIYTKNTTTPKPVFVWLNAEQYASTTTPVFSYKKIVEENIVFVPLNFRLSIFGFICLGVPDAPGNAGLKDILQGLTWLKSNIAGFGGDPNNIVLIGHGSGAALVDLLTMSPRSKNLVHKAIALSGSALSPWAVAYEPVRYAEVFGGVLGITQKSREQLAKLLQTTNPNVLATALEEFKFTNNSLIFAPCIESQNSDPNETIITDAPLNILRSGNYNHVPYIAGFTTREGTLRASEVVFNRWLEGMEANFTNFLPVDLDTNGNATAVQDIKRFYFGDRTIDMGTIEDYLEYQGDTLILLAVIRTARERGVTSTSPVRLLEFGYRGTLNSDWPFNQIPLHGVKHGGFLNYLFDYDLRPSDVQAMNSLVRRLTRFAHTGEPANSAAANVVWSAVDKANLNYLYYGGNDGTGSVAYVEEDRVNPHTTRMTFWNSTYERYYVPPRPVSGANSLLGVALFVTLSQLFFLMF
Summary
Similarity
Belongs to the type-B carboxylesterase/lipase family.
Feature
chain Carboxylic ester hydrolase
Uniprot
H9IX58
A0A0L7LDE4
A0A2S0D925
A0A2H1WJS1
A0A1U9X1V8
A0A212FDJ0
+ More
A0A194QW56 A0A194QEV9 A0A194QF85 A0A291P137 D5G3D9 A0A194QX52 A0A2A4J2J0 A0A1L8D6K3 A0A0C5CGY5 A0A0B4RXS2 B3GEF7 A0A194PFJ7 A0A194QV43 A0A2A4JJU8 A0A2H1VUM3 A0A1L8D6M3 A0A0L7KMN7 A0A1U9X1Y3 A0A1U9X1W8 A0A1U9X1X1 A0A1U9X1U8 A0A2A4J0U9 A0A2H1V317 A0A194RI11 A0A194PHG4 A0A2W1C2P3 A0A2H1WW00 A0A0K8TV95 A0A2H1V568 A0A2A4J321 A0A076FRK9 A0A2H1V340 A0A2H1V381 A0A1U9X1W9 A0A1U9X1S9 A0A2A4J4E2 A0A194PQT2 A0A2H1V5A3 A0A0Y0C087 A0A2A4J4R6 A0A2H1V567 H9JDE9 A0A194QY52 B3TNN2 A0A0F7QEE5 A0A1L8D6K1 H9JDF0 A0A1J0M1C8 A0A2W1BYR0 D9IV62 A0A2H1WVG2 D5G3C9 G1C2I4 A0A0C5BXH0 A0A1J0M1W7 M9WMP0 D3GDM5 A0A0B4RY77 A0A194RGS9 A0A194PP48 A0A2H1WRR7 Q86MC7 A0A1L8D6M6 A0A2A4J4U9 G3CM70 S4WMC9 A0A0K8TV97 D3GDL5 A0A3S2PKB4 A0A2A4J3H7 A0A2H1V569 A0A2A4JGH7 D3GDN1 E5KG19 A0A1I8PWQ3 A0A1I8PWP1 A0A1I8PWQ1 Q171Y4 A0A2H1WXB4 S4WFZ6 A0A127KQ69 A0A3S2LHR9 A0A2H1UZS7 A0A0K0XRW2 A0A1I8PWR0 A0A2H1V6G1 A0A194PL79 A0A0C5C1L6 D3GDL8 A0A2W1BUD0 A4UA25 A0A0U3BZB1
A0A194QW56 A0A194QEV9 A0A194QF85 A0A291P137 D5G3D9 A0A194QX52 A0A2A4J2J0 A0A1L8D6K3 A0A0C5CGY5 A0A0B4RXS2 B3GEF7 A0A194PFJ7 A0A194QV43 A0A2A4JJU8 A0A2H1VUM3 A0A1L8D6M3 A0A0L7KMN7 A0A1U9X1Y3 A0A1U9X1W8 A0A1U9X1X1 A0A1U9X1U8 A0A2A4J0U9 A0A2H1V317 A0A194RI11 A0A194PHG4 A0A2W1C2P3 A0A2H1WW00 A0A0K8TV95 A0A2H1V568 A0A2A4J321 A0A076FRK9 A0A2H1V340 A0A2H1V381 A0A1U9X1W9 A0A1U9X1S9 A0A2A4J4E2 A0A194PQT2 A0A2H1V5A3 A0A0Y0C087 A0A2A4J4R6 A0A2H1V567 H9JDE9 A0A194QY52 B3TNN2 A0A0F7QEE5 A0A1L8D6K1 H9JDF0 A0A1J0M1C8 A0A2W1BYR0 D9IV62 A0A2H1WVG2 D5G3C9 G1C2I4 A0A0C5BXH0 A0A1J0M1W7 M9WMP0 D3GDM5 A0A0B4RY77 A0A194RGS9 A0A194PP48 A0A2H1WRR7 Q86MC7 A0A1L8D6M6 A0A2A4J4U9 G3CM70 S4WMC9 A0A0K8TV97 D3GDL5 A0A3S2PKB4 A0A2A4J3H7 A0A2H1V569 A0A2A4JGH7 D3GDN1 E5KG19 A0A1I8PWQ3 A0A1I8PWP1 A0A1I8PWQ1 Q171Y4 A0A2H1WXB4 S4WFZ6 A0A127KQ69 A0A3S2LHR9 A0A2H1UZS7 A0A0K0XRW2 A0A1I8PWR0 A0A2H1V6G1 A0A194PL79 A0A0C5C1L6 D3GDL8 A0A2W1BUD0 A4UA25 A0A0U3BZB1
EC Number
3.1.1.-
Pubmed
EMBL
BABH01018297
KC200272
AGG20207.1
JTDY01001568
KOB73502.1
KX015848
+ More
ARM65377.1 ODYU01009112 SOQ53299.1 KY021852 AQY62741.1 AGBW02009050 OWR51794.1 KQ461073 KPJ09539.1 KQ459053 KPJ04073.1 KPJ04074.1 MF687634 ATJ44560.1 FJ997303 ADF43468.1 KPJ09540.1 NWSH01003710 PCG65898.1 GEYN01000047 JAV02082.1 KP001157 AJN91202.1 KJ023207 AIY69035.1 BABH01014414 EU688969 ACD80423.1 KQ459606 KPI91798.1 KQ461103 KPJ09403.1 NWSH01001213 PCG72099.1 ODYU01004540 SOQ44535.1 GEYN01000038 JAV02091.1 JTDY01008799 KOB64390.1 KY021872 AQY62761.1 KY021866 AQY62755.1 KY021862 AQY62751.1 KY021846 AQY62735.1 NWSH01003909 PCG65705.1 ODYU01000447 SOQ35238.1 KQ460205 KPJ17074.1 KQ459604 KPI92154.1 KZ149903 PZC78413.1 ODYU01011476 SOQ57231.1 GCVX01000044 JAI18186.1 ODYU01000738 SOQ35977.1 NWSH01003330 PCG66537.1 KF960776 AII21978.1 SOQ35241.1 ODYU01000269 SOQ34744.1 KY021864 AQY62753.1 KY021822 AQY62711.1 PCG66538.1 KQ459602 KPI93490.1 SOQ35979.1 KU215372 AMB19666.1 NWSH01003373 PCG66430.1 SOQ35981.1 BABH01030066 KQ460949 KPJ10397.1 EU339106 ACA50924.1 LC017771 BAR64785.1 GEYN01000046 JAV02083.1 BABH01030065 KU375433 APD13875.1 KZ149915 PZC77960.1 HM191472 MF687569 ADJ96632.1 ATJ44495.1 ODYU01011357 SOQ57053.1 FJ997293 ADF43458.1 JF728803 AEJ38205.1 KP001153 AJN91198.1 KU375436 APD13878.1 KC493657 AGJ94046.1 FJ652457 ACV60241.1 KJ023201 AIY69029.1 KPJ16792.1 KQ459598 KPI94519.1 ODYU01010555 SOQ55751.1 AY091503 AAM14415.1 GEYN01000031 JAV02098.1 NWSH01003365 PCG66460.1 HQ122616 AEL33699.1 KC589105 AGO98723.1 GCVX01000022 JAI18208.1 FJ652447 ACV60231.1 RSAL01000009 RVE53879.1 PCG66541.1 SOQ35983.1 NWSH01001662 PCG70492.1 FJ652463 ACV60247.1 HQ116562 ADR64703.1 CH477443 EAT40802.2 SOQ57054.1 KC589104 AGO98722.1 KT345938 AMO44418.1 RSAL01000115 RVE46908.1 ODYU01000026 SOQ34100.1 KP938880 AKS40361.1 SOQ35982.1 KPI93489.1 KP001156 AJN91201.1 FJ652450 ACV60234.1 PZC77961.1 DQ680828 ABH01081.1 KT345935 ALT14583.1
ARM65377.1 ODYU01009112 SOQ53299.1 KY021852 AQY62741.1 AGBW02009050 OWR51794.1 KQ461073 KPJ09539.1 KQ459053 KPJ04073.1 KPJ04074.1 MF687634 ATJ44560.1 FJ997303 ADF43468.1 KPJ09540.1 NWSH01003710 PCG65898.1 GEYN01000047 JAV02082.1 KP001157 AJN91202.1 KJ023207 AIY69035.1 BABH01014414 EU688969 ACD80423.1 KQ459606 KPI91798.1 KQ461103 KPJ09403.1 NWSH01001213 PCG72099.1 ODYU01004540 SOQ44535.1 GEYN01000038 JAV02091.1 JTDY01008799 KOB64390.1 KY021872 AQY62761.1 KY021866 AQY62755.1 KY021862 AQY62751.1 KY021846 AQY62735.1 NWSH01003909 PCG65705.1 ODYU01000447 SOQ35238.1 KQ460205 KPJ17074.1 KQ459604 KPI92154.1 KZ149903 PZC78413.1 ODYU01011476 SOQ57231.1 GCVX01000044 JAI18186.1 ODYU01000738 SOQ35977.1 NWSH01003330 PCG66537.1 KF960776 AII21978.1 SOQ35241.1 ODYU01000269 SOQ34744.1 KY021864 AQY62753.1 KY021822 AQY62711.1 PCG66538.1 KQ459602 KPI93490.1 SOQ35979.1 KU215372 AMB19666.1 NWSH01003373 PCG66430.1 SOQ35981.1 BABH01030066 KQ460949 KPJ10397.1 EU339106 ACA50924.1 LC017771 BAR64785.1 GEYN01000046 JAV02083.1 BABH01030065 KU375433 APD13875.1 KZ149915 PZC77960.1 HM191472 MF687569 ADJ96632.1 ATJ44495.1 ODYU01011357 SOQ57053.1 FJ997293 ADF43458.1 JF728803 AEJ38205.1 KP001153 AJN91198.1 KU375436 APD13878.1 KC493657 AGJ94046.1 FJ652457 ACV60241.1 KJ023201 AIY69029.1 KPJ16792.1 KQ459598 KPI94519.1 ODYU01010555 SOQ55751.1 AY091503 AAM14415.1 GEYN01000031 JAV02098.1 NWSH01003365 PCG66460.1 HQ122616 AEL33699.1 KC589105 AGO98723.1 GCVX01000022 JAI18208.1 FJ652447 ACV60231.1 RSAL01000009 RVE53879.1 PCG66541.1 SOQ35983.1 NWSH01001662 PCG70492.1 FJ652463 ACV60247.1 HQ116562 ADR64703.1 CH477443 EAT40802.2 SOQ57054.1 KC589104 AGO98722.1 KT345938 AMO44418.1 RSAL01000115 RVE46908.1 ODYU01000026 SOQ34100.1 KP938880 AKS40361.1 SOQ35982.1 KPI93489.1 KP001156 AJN91201.1 FJ652450 ACV60234.1 PZC77961.1 DQ680828 ABH01081.1 KT345935 ALT14583.1
Proteomes
Pfam
PF00135 COesterase
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9IX58
A0A0L7LDE4
A0A2S0D925
A0A2H1WJS1
A0A1U9X1V8
A0A212FDJ0
+ More
A0A194QW56 A0A194QEV9 A0A194QF85 A0A291P137 D5G3D9 A0A194QX52 A0A2A4J2J0 A0A1L8D6K3 A0A0C5CGY5 A0A0B4RXS2 B3GEF7 A0A194PFJ7 A0A194QV43 A0A2A4JJU8 A0A2H1VUM3 A0A1L8D6M3 A0A0L7KMN7 A0A1U9X1Y3 A0A1U9X1W8 A0A1U9X1X1 A0A1U9X1U8 A0A2A4J0U9 A0A2H1V317 A0A194RI11 A0A194PHG4 A0A2W1C2P3 A0A2H1WW00 A0A0K8TV95 A0A2H1V568 A0A2A4J321 A0A076FRK9 A0A2H1V340 A0A2H1V381 A0A1U9X1W9 A0A1U9X1S9 A0A2A4J4E2 A0A194PQT2 A0A2H1V5A3 A0A0Y0C087 A0A2A4J4R6 A0A2H1V567 H9JDE9 A0A194QY52 B3TNN2 A0A0F7QEE5 A0A1L8D6K1 H9JDF0 A0A1J0M1C8 A0A2W1BYR0 D9IV62 A0A2H1WVG2 D5G3C9 G1C2I4 A0A0C5BXH0 A0A1J0M1W7 M9WMP0 D3GDM5 A0A0B4RY77 A0A194RGS9 A0A194PP48 A0A2H1WRR7 Q86MC7 A0A1L8D6M6 A0A2A4J4U9 G3CM70 S4WMC9 A0A0K8TV97 D3GDL5 A0A3S2PKB4 A0A2A4J3H7 A0A2H1V569 A0A2A4JGH7 D3GDN1 E5KG19 A0A1I8PWQ3 A0A1I8PWP1 A0A1I8PWQ1 Q171Y4 A0A2H1WXB4 S4WFZ6 A0A127KQ69 A0A3S2LHR9 A0A2H1UZS7 A0A0K0XRW2 A0A1I8PWR0 A0A2H1V6G1 A0A194PL79 A0A0C5C1L6 D3GDL8 A0A2W1BUD0 A4UA25 A0A0U3BZB1
A0A194QW56 A0A194QEV9 A0A194QF85 A0A291P137 D5G3D9 A0A194QX52 A0A2A4J2J0 A0A1L8D6K3 A0A0C5CGY5 A0A0B4RXS2 B3GEF7 A0A194PFJ7 A0A194QV43 A0A2A4JJU8 A0A2H1VUM3 A0A1L8D6M3 A0A0L7KMN7 A0A1U9X1Y3 A0A1U9X1W8 A0A1U9X1X1 A0A1U9X1U8 A0A2A4J0U9 A0A2H1V317 A0A194RI11 A0A194PHG4 A0A2W1C2P3 A0A2H1WW00 A0A0K8TV95 A0A2H1V568 A0A2A4J321 A0A076FRK9 A0A2H1V340 A0A2H1V381 A0A1U9X1W9 A0A1U9X1S9 A0A2A4J4E2 A0A194PQT2 A0A2H1V5A3 A0A0Y0C087 A0A2A4J4R6 A0A2H1V567 H9JDE9 A0A194QY52 B3TNN2 A0A0F7QEE5 A0A1L8D6K1 H9JDF0 A0A1J0M1C8 A0A2W1BYR0 D9IV62 A0A2H1WVG2 D5G3C9 G1C2I4 A0A0C5BXH0 A0A1J0M1W7 M9WMP0 D3GDM5 A0A0B4RY77 A0A194RGS9 A0A194PP48 A0A2H1WRR7 Q86MC7 A0A1L8D6M6 A0A2A4J4U9 G3CM70 S4WMC9 A0A0K8TV97 D3GDL5 A0A3S2PKB4 A0A2A4J3H7 A0A2H1V569 A0A2A4JGH7 D3GDN1 E5KG19 A0A1I8PWQ3 A0A1I8PWP1 A0A1I8PWQ1 Q171Y4 A0A2H1WXB4 S4WFZ6 A0A127KQ69 A0A3S2LHR9 A0A2H1UZS7 A0A0K0XRW2 A0A1I8PWR0 A0A2H1V6G1 A0A194PL79 A0A0C5C1L6 D3GDL8 A0A2W1BUD0 A4UA25 A0A0U3BZB1
PDB
5THM
E-value=3.00723e-39,
Score=408
Ontologies
Topology
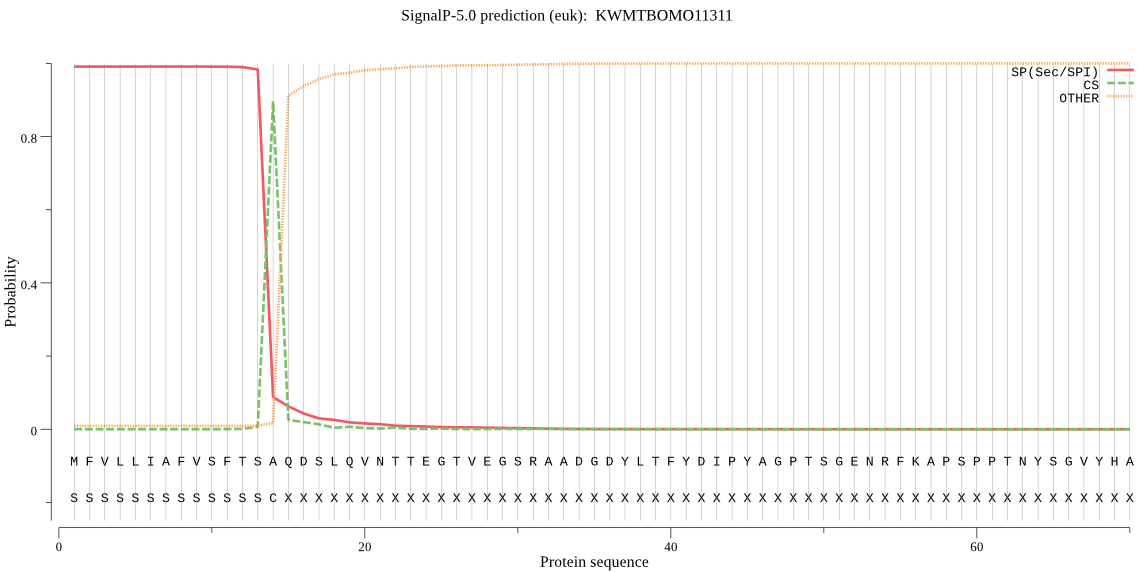
SignalP
Position: 1 - 14,
Likelihood: 0.990928
Length:
559
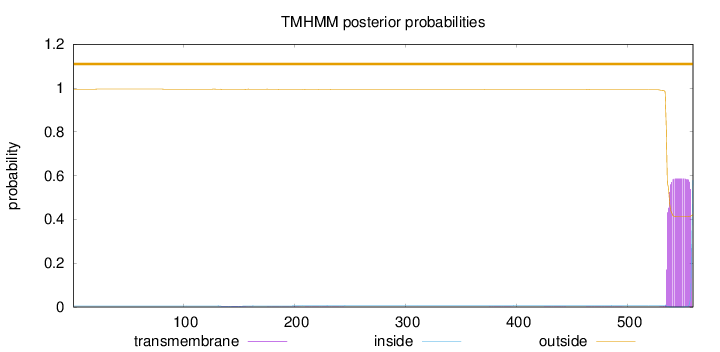
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
13.03422
Exp number, first 60 AAs:
0.00092
Total prob of N-in:
0.00499
outside
1 - 559
Population Genetic Test Statistics
Pi
227.572787
Theta
182.252282
Tajima's D
0.503834
CLR
0.385681
CSRT
0.512224388780561
Interpretation
Uncertain
