Gene
KWMTBOMO11306
Pre Gene Modal
BGIBMGA001988
Annotation
PREDICTED:_transient_receptor_potential_cation_channel_trpm_isoform_X2_[Bombyx_mori]
Full name
Transient receptor potential cation channel trpm
Alternative Name
Transient receptor potential cation channel, subfamily M ortholog
Location in the cell
PlasmaMembrane Reliability : 4.333
Sequence
CDS
ATGCTCGCGGAGAATCAGGAGAGAGAAGAGAGAACTTGCGACACGCATTCGTCGACGCCCGTCGTGTGTCTGGTCATCGAAGGTGGAACGAACACAGTGAGAGCGGTTCTGGAGTACGTGACTGATAACCCACCGGTGCCGGTGGTGGTCTGCGATGGAAGCGGTCGGGCAGCGGACCTCATCGCTTTTGTTCATAAGTACGCATCGGAAACCGGCGAACAAACAGTGCTGGAGTCAATGCGAGATTACCTGATAGGCACCATACAGAAGACCTTCGAAGTCGGACTGCAGCAAGCTGAGTGTTTGTACACGGAACTATTGCAGTGCACGAGAAAGAAGAATTTGATAACGGTCTTCAGGATTCAAGAGCGTGCAGAAGGTGGAGAAGTCCAAGAGCTTGATCAGGTCATACTGACGGCGTTGTTCCGGGCCCAGCATCTGACTCCAAGTGAACAGTTGTCTTTAGCCCTCACTTGGAACCGGGTCGACATCGCCAGATCCGAGATCTTCGTTTACGGACAGGAATGGCCTCCAGGCGCCCTAGACGAAGCCATGATGCAAGCCCTGGAGCACGATAGGATCGACTTCGTTAAGCTTCTCCTCGAGAACGGCGTGTCCATGAGGAAATTCCTCACGATTCCTAGACTTGAAGAACTTTACAACACGAAGAGCGGGCCTAGTAACACCTTGAGGTACATCTTAAGGGACGTGAGGCCTCATCTGCCCAAAGGCTACGTGTACACGCTTCACGACATCGGTTTGGTGATCAACAAACTCATTGGCGGAGCTTATAGGTGCTACTACACACGTCGGAAGTTTCGACCGATCTACGCAAAGGTGATGAACAAAAGCGTCAATGTACATCGCAACAGTGCGTCCTTCACCAGGCATAACGCTGGAGGCCTGTCGCTCATCACCGGGTTCATGCCGGTCACCAGTGAAATGGCGCTTTTCGATTATCCTTTCAACGAGCTTTTAATGTGGGCAGTCCTAACGAAGAGACATCAAATGGCGCTCCTCATGTGGACACATGGCGAGGAGTCGCTTGCGAAATCGCTCATAGCATGCAAGCTGTACAAGGCCATGGCCCACGAGGCAGCCGAGGATGATATGGAGACTGAAGTTTATGAAGAGCTAAGGCACTACGGCAAAGAATTTGAGAATAAGGCTTTGGAGCTTCTAGATTACTGCTACCGCCAAGATGATGATCAGGCCCAGCAACTCCTCACGTGCGAGCTCCAGAACTGGTCCGGTCAGACCTGTCTCAGCTTAGCCGTGACCGCGAACCATCGTGCGCTACTCGCTCATCCGTGCTCACAAATAATCTTAGCGGACTTATGGATGGGCGGCCTCAGAACGAGAAAGAACACCAATCTCAAGGTAATTCTGAGCCTGCTGTGTCCGCTGTACATTCTGAGATTGGAGTTCAAGTCCAAAGAGGAACTTCAGCTGATGCCGCAGACGGAAGAAGAGCATCTAGAGAACGAGAGCATCGAGGATGATAGGAGCACCAAGGATCCGACAGATGCCGAGCAGGCGTTGATTGGCTCAGGGGAATGCGAGACCCGGGTCGATCAGAACGGGAAAGTTGGTAAAGTAGGATGGGAACCATCGTACAGAGAGCTGCCAGAGTACCACCCATCTGAAATTAAACGAATGAGGCCACTGCGACTGAGGAAGAAGATCTATGAGTTCTTTACGGCACCGATCACTAAATTTTGGGCCGATTCGATAGCCTACATATTGTTCCTGTTGATGTTTACATACACAGTATTAGTCAAGATGAATCCTACACCTTCTTGGCCGGAAATATATTCTATATGTTATATATTAACTTTCTTATGCGAGAAGATTAGAGAAATCGTAACATCCGAGCCTGTTGCCATTAGACATAAGTTCAGTGTGTGGGCATGGAACATGTGGAACACGTATGACGCTGGTTTTATAATATTCTTCTTAGTTGGATTGACGCTGAGACTCAGAGAGATCTCAAGAGACGTAGGGAGAGTTATATACTGCGTCGATATTATATATTGGTACCTAAGGATCTTGAATATATTGGGTGTCAATAAGTACTTAGGTCCCCTAGTCACAATGATGGGTAAAATGGTAAAGAACATGATATACTTCGTGGTGCTATTGCTAGTGGTATTAATGTCTTTCGGTGTTGCCAGACAAGCTATACTCCATCCAGATAAGGACGCTAGTTGGCATCTAATCAGAGAGGTTTTCTTCCAACCGTACTTCATGCTCTATGGGGAAGTATTCGCCGAGCAGATGGCGCCTCCTTGTGGCCTTCCGGGCCTTGAGGAGTGCCGGGTCGGCCACTGGGTCCCCCCGCTCGTCATGAGCGTATATCTGCTCATCGCTAACATATTACTCGTCAATCTTCTAATAGCCGTCTTCAATAACATCTTTATAGAAGTCAACGGAGTTTCCCATCAGGTTTCGATCGCGTTCATTACGTAG
Protein
MLAENQEREERTCDTHSSTPVVCLVIEGGTNTVRAVLEYVTDNPPVPVVVCDGSGRAADLIAFVHKYASETGEQTVLESMRDYLIGTIQKTFEVGLQQAECLYTELLQCTRKKNLITVFRIQERAEGGEVQELDQVILTALFRAQHLTPSEQLSLALTWNRVDIARSEIFVYGQEWPPGALDEAMMQALEHDRIDFVKLLLENGVSMRKFLTIPRLEELYNTKSGPSNTLRYILRDVRPHLPKGYVYTLHDIGLVINKLIGGAYRCYYTRRKFRPIYAKVMNKSVNVHRNSASFTRHNAGGLSLITGFMPVTSEMALFDYPFNELLMWAVLTKRHQMALLMWTHGEESLAKSLIACKLYKAMAHEAAEDDMETEVYEELRHYGKEFENKALELLDYCYRQDDDQAQQLLTCELQNWSGQTCLSLAVTANHRALLAHPCSQIILADLWMGGLRTRKNTNLKVILSLLCPLYILRLEFKSKEELQLMPQTEEEHLENESIEDDRSTKDPTDAEQALIGSGECETRVDQNGKVGKVGWEPSYRELPEYHPSEIKRMRPLRLRKKIYEFFTAPITKFWADSIAYILFLLMFTYTVLVKMNPTPSWPEIYSICYILTFLCEKIREIVTSEPVAIRHKFSVWAWNMWNTYDAGFIIFFLVGLTLRLREISRDVGRVIYCVDIIYWYLRILNILGVNKYLGPLVTMMGKMVKNMIYFVVLLLVVLMSFGVARQAILHPDKDASWHLIREVFFQPYFMLYGEVFAEQMAPPCGLPGLEECRVGHWVPPLVMSVYLLIANILLVNLLIAVFNNIFIEVNGVSHQVSIAFIT
Summary
Description
Calcium channel mediating constitutive calcium ion entry.
Similarity
Belongs to the transient receptor (TC 1.A.4) family. LTrpC subfamily.
Keywords
Alternative splicing
Complete proteome
Ion channel
Ion transport
Membrane
Phosphoprotein
Reference proteome
RNA editing
Transmembrane
Transmembrane helix
Transport
Feature
chain Transient receptor potential cation channel trpm
splice variant In isoform B, isoform C and isoform D.
splice variant In isoform B, isoform C and isoform D.
Uniprot
A0A194R122
A0A3S2P3F4
H9IXK5
A0A084WMT1
A0A182HAI6
A0A182RUF8
+ More
A0A1S4G7E1 A0A1L8DKF9 A0A182N9J3 A0A182WB94 A0A182X2A3 A0A1I8M2P8 W4VRK0 A0A384T281 A0A1I8JT75 Q7PT99 Q16JC7 A0A0A1WNF2 A0A182FFL8 A0A067R873 A0A2M4A299 A0A2M4A2H3 A0A2M4A2E1 A0A2M4B8G9 A0A0K8WAM8 W8AZI2 A0A2M4CSI3 W8AKE0 A0A1I8M2P6 A0A182PIT3 A0A0A1XCB9 A0A1B6DW32 A0A0K8UMW6 A0A1B6CHW9 A0A1I8M2Q3 A0A2M4CKX6 A0A1I8M2Q0 A0A0K8W6A0 A0A1I8M2P5 A0A0R3NKP2 A0A1Y1MEE7 A0A0P8Y3Q1 A0A1I8M2Q1 A0A034V3S7 A0A0P9BXU3 A0A0R3NKT9 A0A0R3NKI6 A0A0Q9XGL4 A0A0Q9XJP9 A0A2M3Z0A8 A0A1I8M2P9 A0A1I8M2P7 A0A0P9AEA7 A0A0Q9VY26 A0A0Q9W722 A0A3B0J3H7 A0A0R1DR22 A0A2M4CKI8 A0A0Q9XIH8 A0A0J9RD37 A0A034V838 A0A0R1DT00 A0A0Q5VMK0 A0A0Q5VLM6 A0A0Q9W9P5 A0A1I8M2Q2 A0A1B6E5Z4 B4MFT5 A0A0R1DWM3 A0A0Q5VMF2 A0A0Q9W5Y3 A0A2M4B8H7 A0A0J9RD88 A0A0Q9W945 A0A0Q9X7N1 B3NQF3 A0A1W4WC30 A0A0J9RDE2 B4P890 A0A1W4W0D3 A0A1W4WCH8 A0A0J9REF0 A0A2M4CSJ8 A0A1W4W0E0 A0A0Q9VY01 A0A0Q9X9I3 A0A1W4WB55 A0A0Q9VYT9 A0A0Q9VY23 B4MJT0 A0A1I8P4R3 A0A0Q9VY84 A0A0Q9X0W9 A8DYE2 A0A0Q9WAC5 A0A0Q5VNM3 A0A1W4WCF7 B7YZH3 A0A0Q5W046 A0A0R1DUZ6
A0A1S4G7E1 A0A1L8DKF9 A0A182N9J3 A0A182WB94 A0A182X2A3 A0A1I8M2P8 W4VRK0 A0A384T281 A0A1I8JT75 Q7PT99 Q16JC7 A0A0A1WNF2 A0A182FFL8 A0A067R873 A0A2M4A299 A0A2M4A2H3 A0A2M4A2E1 A0A2M4B8G9 A0A0K8WAM8 W8AZI2 A0A2M4CSI3 W8AKE0 A0A1I8M2P6 A0A182PIT3 A0A0A1XCB9 A0A1B6DW32 A0A0K8UMW6 A0A1B6CHW9 A0A1I8M2Q3 A0A2M4CKX6 A0A1I8M2Q0 A0A0K8W6A0 A0A1I8M2P5 A0A0R3NKP2 A0A1Y1MEE7 A0A0P8Y3Q1 A0A1I8M2Q1 A0A034V3S7 A0A0P9BXU3 A0A0R3NKT9 A0A0R3NKI6 A0A0Q9XGL4 A0A0Q9XJP9 A0A2M3Z0A8 A0A1I8M2P9 A0A1I8M2P7 A0A0P9AEA7 A0A0Q9VY26 A0A0Q9W722 A0A3B0J3H7 A0A0R1DR22 A0A2M4CKI8 A0A0Q9XIH8 A0A0J9RD37 A0A034V838 A0A0R1DT00 A0A0Q5VMK0 A0A0Q5VLM6 A0A0Q9W9P5 A0A1I8M2Q2 A0A1B6E5Z4 B4MFT5 A0A0R1DWM3 A0A0Q5VMF2 A0A0Q9W5Y3 A0A2M4B8H7 A0A0J9RD88 A0A0Q9W945 A0A0Q9X7N1 B3NQF3 A0A1W4WC30 A0A0J9RDE2 B4P890 A0A1W4W0D3 A0A1W4WCH8 A0A0J9REF0 A0A2M4CSJ8 A0A1W4W0E0 A0A0Q9VY01 A0A0Q9X9I3 A0A1W4WB55 A0A0Q9VYT9 A0A0Q9VY23 B4MJT0 A0A1I8P4R3 A0A0Q9VY84 A0A0Q9X0W9 A8DYE2 A0A0Q9WAC5 A0A0Q5VNM3 A0A1W4WCF7 B7YZH3 A0A0Q5W046 A0A0R1DUZ6
Pubmed
EMBL
KQ461073
KPJ09541.1
RSAL01000034
RVE51394.1
BABH01018315
BABH01018316
+ More
BABH01018317 BABH01018318 ATLV01024507 ATLV01024508 ATLV01024509 KE525352 KFB51525.1 JXUM01005304 JXUM01005305 JXUM01005306 JXUM01005307 JXUM01005308 JXUM01005309 JXUM01005310 KQ560190 KXJ83914.1 GFDF01007254 JAV06830.1 GANO01002328 JAB57543.1 KX249699 AOR81476.1 APCN01003636 AAAB01008807 EAA04234.4 CH478017 EAT34369.1 GBXI01013708 JAD00584.1 KK852639 KDR19648.1 GGFK01001554 MBW34875.1 GGFK01001517 MBW34838.1 GGFK01001569 MBW34890.1 GGFJ01000151 MBW49292.1 GDHF01004167 JAI48147.1 GAMC01020269 JAB86286.1 GGFL01004112 MBW68290.1 GAMC01020268 JAB86287.1 GBXI01005697 JAD08595.1 GEDC01007412 JAS29886.1 GDHF01024393 JAI27921.1 GEDC01024308 JAS12990.1 GGFL01001390 MBW65568.1 GDHF01005914 JAI46400.1 CM000071 KRT01474.1 GEZM01035131 GEZM01035128 JAV83388.1 CH902619 KPU76305.1 KPU76306.1 KPU76308.1 GAKP01021006 JAC37946.1 KPU76304.1 KRT01476.1 KRT01464.1 CH933808 KRG04098.1 KRG04088.1 GGFM01001209 MBW21960.1 KPU76309.1 CH940667 KRF77706.1 KRF77697.1 OUUW01000001 SPP75907.1 CM000158 KRJ99640.1 GGFL01001601 MBW65779.1 KRG04087.1 CM002911 KMY93948.1 GAKP01021007 JAC37945.1 KRJ99645.1 CH954179 KQS62561.1 KQS62557.1 KRF77695.1 GEDC01003984 JAS33314.1 EDW57256.2 KRJ99648.1 KQS62563.1 KRF77703.1 GGFJ01000195 MBW49336.1 KMY93937.1 KRF77700.1 KRG04093.1 EDV55929.2 KMY93951.1 EDW91130.2 KMY93939.1 GGFL01004119 MBW68297.1 KRF77705.1 KRG04089.1 KRF77702.1 KRF77696.1 KRF77698.1 KRF77699.1 CH963846 EDW72369.2 KRF77704.1 KRF97549.1 AE013599 AY069181 KRF77694.1 KQS62556.1 ACL83127.1 KQS62562.1 KRJ99650.1
BABH01018317 BABH01018318 ATLV01024507 ATLV01024508 ATLV01024509 KE525352 KFB51525.1 JXUM01005304 JXUM01005305 JXUM01005306 JXUM01005307 JXUM01005308 JXUM01005309 JXUM01005310 KQ560190 KXJ83914.1 GFDF01007254 JAV06830.1 GANO01002328 JAB57543.1 KX249699 AOR81476.1 APCN01003636 AAAB01008807 EAA04234.4 CH478017 EAT34369.1 GBXI01013708 JAD00584.1 KK852639 KDR19648.1 GGFK01001554 MBW34875.1 GGFK01001517 MBW34838.1 GGFK01001569 MBW34890.1 GGFJ01000151 MBW49292.1 GDHF01004167 JAI48147.1 GAMC01020269 JAB86286.1 GGFL01004112 MBW68290.1 GAMC01020268 JAB86287.1 GBXI01005697 JAD08595.1 GEDC01007412 JAS29886.1 GDHF01024393 JAI27921.1 GEDC01024308 JAS12990.1 GGFL01001390 MBW65568.1 GDHF01005914 JAI46400.1 CM000071 KRT01474.1 GEZM01035131 GEZM01035128 JAV83388.1 CH902619 KPU76305.1 KPU76306.1 KPU76308.1 GAKP01021006 JAC37946.1 KPU76304.1 KRT01476.1 KRT01464.1 CH933808 KRG04098.1 KRG04088.1 GGFM01001209 MBW21960.1 KPU76309.1 CH940667 KRF77706.1 KRF77697.1 OUUW01000001 SPP75907.1 CM000158 KRJ99640.1 GGFL01001601 MBW65779.1 KRG04087.1 CM002911 KMY93948.1 GAKP01021007 JAC37945.1 KRJ99645.1 CH954179 KQS62561.1 KQS62557.1 KRF77695.1 GEDC01003984 JAS33314.1 EDW57256.2 KRJ99648.1 KQS62563.1 KRF77703.1 GGFJ01000195 MBW49336.1 KMY93937.1 KRF77700.1 KRG04093.1 EDV55929.2 KMY93951.1 EDW91130.2 KMY93939.1 GGFL01004119 MBW68297.1 KRF77705.1 KRG04089.1 KRF77702.1 KRF77696.1 KRF77698.1 KRF77699.1 CH963846 EDW72369.2 KRF77704.1 KRF97549.1 AE013599 AY069181 KRF77694.1 KQS62556.1 ACL83127.1 KQS62562.1 KRJ99650.1
Proteomes
UP000053240
UP000283053
UP000005204
UP000030765
UP000069940
UP000249989
+ More
UP000075900 UP000075884 UP000075920 UP000076407 UP000095301 UP000075840 UP000007062 UP000008820 UP000069272 UP000027135 UP000075885 UP000001819 UP000007801 UP000009192 UP000008792 UP000268350 UP000002282 UP000008711 UP000192221 UP000007798 UP000095300 UP000000803
UP000075900 UP000075884 UP000075920 UP000076407 UP000095301 UP000075840 UP000007062 UP000008820 UP000069272 UP000027135 UP000075885 UP000001819 UP000007801 UP000009192 UP000008792 UP000268350 UP000002282 UP000008711 UP000192221 UP000007798 UP000095300 UP000000803
Interpro
Gene 3D
ProteinModelPortal
A0A194R122
A0A3S2P3F4
H9IXK5
A0A084WMT1
A0A182HAI6
A0A182RUF8
+ More
A0A1S4G7E1 A0A1L8DKF9 A0A182N9J3 A0A182WB94 A0A182X2A3 A0A1I8M2P8 W4VRK0 A0A384T281 A0A1I8JT75 Q7PT99 Q16JC7 A0A0A1WNF2 A0A182FFL8 A0A067R873 A0A2M4A299 A0A2M4A2H3 A0A2M4A2E1 A0A2M4B8G9 A0A0K8WAM8 W8AZI2 A0A2M4CSI3 W8AKE0 A0A1I8M2P6 A0A182PIT3 A0A0A1XCB9 A0A1B6DW32 A0A0K8UMW6 A0A1B6CHW9 A0A1I8M2Q3 A0A2M4CKX6 A0A1I8M2Q0 A0A0K8W6A0 A0A1I8M2P5 A0A0R3NKP2 A0A1Y1MEE7 A0A0P8Y3Q1 A0A1I8M2Q1 A0A034V3S7 A0A0P9BXU3 A0A0R3NKT9 A0A0R3NKI6 A0A0Q9XGL4 A0A0Q9XJP9 A0A2M3Z0A8 A0A1I8M2P9 A0A1I8M2P7 A0A0P9AEA7 A0A0Q9VY26 A0A0Q9W722 A0A3B0J3H7 A0A0R1DR22 A0A2M4CKI8 A0A0Q9XIH8 A0A0J9RD37 A0A034V838 A0A0R1DT00 A0A0Q5VMK0 A0A0Q5VLM6 A0A0Q9W9P5 A0A1I8M2Q2 A0A1B6E5Z4 B4MFT5 A0A0R1DWM3 A0A0Q5VMF2 A0A0Q9W5Y3 A0A2M4B8H7 A0A0J9RD88 A0A0Q9W945 A0A0Q9X7N1 B3NQF3 A0A1W4WC30 A0A0J9RDE2 B4P890 A0A1W4W0D3 A0A1W4WCH8 A0A0J9REF0 A0A2M4CSJ8 A0A1W4W0E0 A0A0Q9VY01 A0A0Q9X9I3 A0A1W4WB55 A0A0Q9VYT9 A0A0Q9VY23 B4MJT0 A0A1I8P4R3 A0A0Q9VY84 A0A0Q9X0W9 A8DYE2 A0A0Q9WAC5 A0A0Q5VNM3 A0A1W4WCF7 B7YZH3 A0A0Q5W046 A0A0R1DUZ6
A0A1S4G7E1 A0A1L8DKF9 A0A182N9J3 A0A182WB94 A0A182X2A3 A0A1I8M2P8 W4VRK0 A0A384T281 A0A1I8JT75 Q7PT99 Q16JC7 A0A0A1WNF2 A0A182FFL8 A0A067R873 A0A2M4A299 A0A2M4A2H3 A0A2M4A2E1 A0A2M4B8G9 A0A0K8WAM8 W8AZI2 A0A2M4CSI3 W8AKE0 A0A1I8M2P6 A0A182PIT3 A0A0A1XCB9 A0A1B6DW32 A0A0K8UMW6 A0A1B6CHW9 A0A1I8M2Q3 A0A2M4CKX6 A0A1I8M2Q0 A0A0K8W6A0 A0A1I8M2P5 A0A0R3NKP2 A0A1Y1MEE7 A0A0P8Y3Q1 A0A1I8M2Q1 A0A034V3S7 A0A0P9BXU3 A0A0R3NKT9 A0A0R3NKI6 A0A0Q9XGL4 A0A0Q9XJP9 A0A2M3Z0A8 A0A1I8M2P9 A0A1I8M2P7 A0A0P9AEA7 A0A0Q9VY26 A0A0Q9W722 A0A3B0J3H7 A0A0R1DR22 A0A2M4CKI8 A0A0Q9XIH8 A0A0J9RD37 A0A034V838 A0A0R1DT00 A0A0Q5VMK0 A0A0Q5VLM6 A0A0Q9W9P5 A0A1I8M2Q2 A0A1B6E5Z4 B4MFT5 A0A0R1DWM3 A0A0Q5VMF2 A0A0Q9W5Y3 A0A2M4B8H7 A0A0J9RD88 A0A0Q9W945 A0A0Q9X7N1 B3NQF3 A0A1W4WC30 A0A0J9RDE2 B4P890 A0A1W4W0D3 A0A1W4WCH8 A0A0J9REF0 A0A2M4CSJ8 A0A1W4W0E0 A0A0Q9VY01 A0A0Q9X9I3 A0A1W4WB55 A0A0Q9VYT9 A0A0Q9VY23 B4MJT0 A0A1I8P4R3 A0A0Q9VY84 A0A0Q9X0W9 A8DYE2 A0A0Q9WAC5 A0A0Q5VNM3 A0A1W4WCF7 B7YZH3 A0A0Q5W046 A0A0R1DUZ6
PDB
5ZX5
E-value=3.02287e-177,
Score=1600
Ontologies
GO
Topology
Subcellular location
Membrane
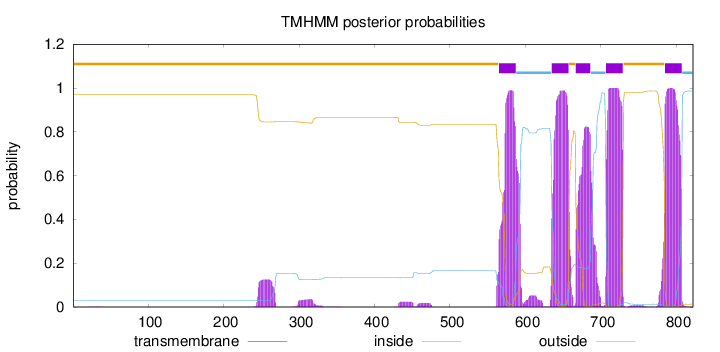
Length:
822
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
110.75458
Exp number, first 60 AAs:
0.00463
Total prob of N-in:
0.03070
outside
1 - 564
TMhelix
565 - 587
inside
588 - 634
TMhelix
635 - 657
outside
658 - 666
TMhelix
667 - 686
inside
687 - 706
TMhelix
707 - 729
outside
730 - 784
TMhelix
785 - 807
inside
808 - 822
Population Genetic Test Statistics
Pi
236.641274
Theta
164.451781
Tajima's D
0.901583
CLR
0.076091
CSRT
0.632818359082046
Interpretation
Uncertain
