Gene
KWMTBOMO11302 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001836
Annotation
PREDICTED:_cartilage_oligomeric_matrix_protein_[Papilio_machaon]
Location in the cell
Extracellular Reliability : 2.144 Nuclear Reliability : 1.708
Sequence
CDS
ATGTCCGGGACTTACGACCTTCTGGATATACTGAGGGACCAGAAGACGAAATTGTATCACGAAGAAAACGCTCCAGTCGAAATATACGGCAGCGAAAAAGCAGCGGTTGAATCTTTGAGTTGTGCAAAAGACAACATCAGACCTCCAACTCTACTGAGACTGGACGAACCTGAATCGGACGTAGAAGAAGTCCAAGAATTCAGAAAGAAGGAAGCTCTCATGGACAGAGAGGAAGAAATGCAAGGAGATGATGCTACTAATTATTTGGAAGCTAACGGTATTCTGCCGCAACCTTCAAGAGGGGATATACCTCAGTCTGCCATTGAATCGTGTGATGATGAAGTCATCCGTCAACTGACCCTACTGCGTCAGATCATCGAGCACTTGCGCCGGGAGGTCGGGGATCAGAAGCACACAATAGACAGCCTAAGGTCTCAACTCCACCAGTGCTGCAACAGAGTACCCGTCACCCCGCCCCCCGTGGAGAGATGCTCCGGATCCTCATGCTACCCTGGGGTCGAATGCCGGAACACTGTCGCGGGGTTCGAGTGTGGGTCGTGTCCGCGCGGCATGGAGGGCGACGGCCGGAGGTGCAGACCCGTACTGTGTAACAGACGCCCATGCTCACAAGGGGAGAGGTGCGTCGATACGGACCAAGGTTTCCGGTGCGAGAGATGCCCAGGGTCACAGACGAGTGATGGCCAGACCTGCAGGACTGCGTGTAGCGCTAACCCTTGCTTTGGAGGACGAGTACCGTGCCAGGACTTACCAGACGGCAGATACCAATGCGGCAAGTGCCCTTCTGGGTTCCACGGGAATGGAGTCGAGTGCTATAGGATATCCTGCAGAGCAAACACGTGCTTTCAAGGTGTGAGCTGCGAGGAGACCCAATATGGTCCTAAATGCGGGTCCTGTCCCACTGGGTACGTGGGAGACGGTCGGCAGTGCCAGCACGTGTGTGACGCTCAGAAGCCCTGCGGTAGAGAGCGACGCTGTATGCCCAAAAGTACCAGCCCTTATTACGAGTGTGAGGGATGTCCCAGCGGCTACCAGTGGAATGGGTACACGTGCGTGGATATGGACGAATGCGATTTGATTCGGCCCTGTGATGATCTGGTCTCGTGTCGCAACGAAGAAGGCGGCTTCACCTGTGGGCCGTGTCCTCCAGGGTTCACAGGTAGCCAGGGCTGGCGGGGCACTGGCAATGAGAGACGAAGGGAACATTGCGTGGATGTGGACGAGTGCGCTGACGGACGAGCCAACTGTCCAAGAGGACGGCTTTGCGTCAATACCCCGGGCTCATACATCTGCGTTCCCTGCGGCGGGCACTACTACGTGAACACGACGAGGCCGTGCTTCGACCCGGACTCGCTGGTGAAGCGGTGCGACCCCAGCTACTGCAGGCGATTCAACGCCGTCTGTGGTTTCGGACAAAAGTGCGTTTGTAATACCGGTTGGACTGGCAACGGTACAGTGTGCGGACTGGACCGAGATCTAGACGGGCATCCGGACGAGCAGCTGCCTTGTAATGAGCCTCGCTGCAAGAAGGACAACTGCCCCGACGTCTCCAACTCCGGGCAAGAGGACGTGGACGGTGACGGCATCGGGGACTCGTGCGATTCTGATGCAGACGGAGACGGGATTCCCAATTTTCCTGACAACTGCCCGTTGACCCCCAACCCGGACCAGTTGGACGACGACGAGAACAAATCGGACGAGAGAGGAAACGCCTGTGACAACTGTCCCCGGCACTACAACCCTCGCCAGGAGGACACCGACGGAGACGGAACGGGAGACGTCTGTGATTCGGACATGGACGATGACGACATACCGAACCAAGTGGACAACTGCCCCAGGAAGTACAATCCGCAGCAAGACGACATGGACTCGGACGGCGTCGGCGACGTCTGCGACAACTGCCCCCGCGTCCGCAACACGAACCAAGAGGACACGGACAAGGACAACGTCGGCGACGCGTGCGACAGCGATATAGACCGTGACCAAGACGGGATACAAGATGGCTTAGACAACTGCCCGCTGATACCGAACAGCGACCAACAGGACGTGGACCGCGACGACAAGGGCGACGTCTGTGACGACGACGACGACGGAGATGGGATACCGGATCTGGACGACAACTGCCAGATCGTGCCCAATAGGGATCAAAAGGACTCTGATAATGACGGCATTGGAGACGCCTGTGACAATGACAATGACGGCGACCGGGTCATCAACGAGCTGGACAACTGCCCGAACAATTCGCAGATATTTCGAACGGACTTCAGACAGTTCACCACAGTCCGATTGGACCCTGAAGGCGAGTCTCAACAGGAGCCCAACTGGAGGGTGACGGACAACGGAGCTGAGATCCTGCAGACCCTGAACTCGGACCCTGGTCTGGCGGTCGGATTCGACAGCTTCGGCGGAGTCGACTACGAGGGGACCATCTTCGTCAACACAAAGATAGATGACGATTATGTTGGATTTATTTTTGGATACCAAAGCAACAAGAGGTTTTATGTGGTCATGTGGAAGAAGGATAAACAGACTTATTGGCAGAAGACTCCGTTCAGAGCGGTCGCCGAGAACGGCATACAGCTCAAGCTGGTCAACTCGAAGACTGGCCCGGGGAAGATACTCAGGAACGCTCTCTGGAATACTAACTCTACCCCGGATCAGGTCACCCTGCTCTGGAAAGACCCTCGCAACGTGGGCTGGCGCGAGCAGACGGCGTACCGCTGGAAGCTCCTCCACCGCCCCAAGATCGGTCTCATCAGACTGAAGATGTACGAGAACAACCGCCTGGTCGCGGACTCTGGGAACACTTACGACTTCACCCTGAAGGGGGGGAGACTTGGAGTGTTCTGCTTCTCCCAGGAAATGATCATATGGTCTAATCTACTCTACAAATGTAATGATAAGATTCCGGCGAGCATAGCATCGGAGATTTCAACAAGGAACCATTTCGAAATCGACCAAGATTATTTCACAAACGAGTAG
Protein
MSGTYDLLDILRDQKTKLYHEENAPVEIYGSEKAAVESLSCAKDNIRPPTLLRLDEPESDVEEVQEFRKKEALMDREEEMQGDDATNYLEANGILPQPSRGDIPQSAIESCDDEVIRQLTLLRQIIEHLRREVGDQKHTIDSLRSQLHQCCNRVPVTPPPVERCSGSSCYPGVECRNTVAGFECGSCPRGMEGDGRRCRPVLCNRRPCSQGERCVDTDQGFRCERCPGSQTSDGQTCRTACSANPCFGGRVPCQDLPDGRYQCGKCPSGFHGNGVECYRISCRANTCFQGVSCEETQYGPKCGSCPTGYVGDGRQCQHVCDAQKPCGRERRCMPKSTSPYYECEGCPSGYQWNGYTCVDMDECDLIRPCDDLVSCRNEEGGFTCGPCPPGFTGSQGWRGTGNERRREHCVDVDECADGRANCPRGRLCVNTPGSYICVPCGGHYYVNTTRPCFDPDSLVKRCDPSYCRRFNAVCGFGQKCVCNTGWTGNGTVCGLDRDLDGHPDEQLPCNEPRCKKDNCPDVSNSGQEDVDGDGIGDSCDSDADGDGIPNFPDNCPLTPNPDQLDDDENKSDERGNACDNCPRHYNPRQEDTDGDGTGDVCDSDMDDDDIPNQVDNCPRKYNPQQDDMDSDGVGDVCDNCPRVRNTNQEDTDKDNVGDACDSDIDRDQDGIQDGLDNCPLIPNSDQQDVDRDDKGDVCDDDDDGDGIPDLDDNCQIVPNRDQKDSDNDGIGDACDNDNDGDRVINELDNCPNNSQIFRTDFRQFTTVRLDPEGESQQEPNWRVTDNGAEILQTLNSDPGLAVGFDSFGGVDYEGTIFVNTKIDDDYVGFIFGYQSNKRFYVVMWKKDKQTYWQKTPFRAVAENGIQLKLVNSKTGPGKILRNALWNTNSTPDQVTLLWKDPRNVGWREQTAYRWKLLHRPKIGLIRLKMYENNRLVADSGNTYDFTLKGGRLGVFCFSQEMIIWSNLLYKCNDKIPASIASEISTRNHFEIDQDYFTNE
Summary
Uniprot
A0A2H1VZK9
A0A194QKU2
A0A0L7LQW3
A0A212EZM8
H9IX53
A0A194QVH8
+ More
A0A3S2TP60 A0A2A4JBX4 A0A2A4JDE5 E0VQ35 A0A1Y1KFW1 A0A1Y1KC59 A0A2J7RKL3 A0A0K8SN26 A0A146LPP8 A0A0A9ZDF3 A0A1B1M0Q9 A0A2H8TD90 J9K964 A0A224XDW2 A0A182FNT9 A0A1S4FIM7 Q16ZU9 B0WVE0 A0A2S2R1N5 W5JW93 A0A1Q3FPU0 A0A023F3J1 A0A182P4E3 A0A182QRY4 A0A084WSA5 Q7QK54 A0A182UXT1 A0A182HZQ6 A0A182NMX8 A0A182U6A2 A0A182RN68 A0A182XWG9 A0A1J1HLI3 A0A182VYF8 A0A182IR04 A0A182MKF1 A0A182XGM1 A0A0K2TRS2 A0A2L2Y9F6 A0A2P2I1C0 A0A3B0JUD1 A0A1B6L9W4 A0A0P4VVB7 B4GJY3 A0A0P4Z3E1 A0A0N8A8K8 A0A0P5S5Z0 A0A162DFK5 A0A0P5FR53 A0A0P5CM97 A0A0N8CAB5 A0A0N8A3Q2 A0A0P5F7N0 A0A0P5XU95 A0A0N8C4G0 A0A0P5EK42 A0A0P5HG21 A0A0P5AS87 A0A0P4ZTS6 A0A1Y1L6J8 A0A0P4YIM1 A0A0P5UTC7 A0A2C9JV62 A0A0N8CUE6 A0A0P4Y347 A0A0P5EI44 A0A0P5CE47 A0A0P5IWS3 A0A0P5C8R0 A0A0P5VR40 A0A0P5CG86 K1PQ45 A0A0N8A7V3 A0A0P5R6U7 A0A0P5CGB0 A0A0N8BSE3 A0A0P5N2P7 A0A0P5XTT6 A0A0P5K353 A0A0P5CZW4 A0A0K8W363 A0A0P4Y4N3 A0A0P5CS41 A0A0P5WL95 A0A0P4Y4B1 A0A0P6B4D3 A0A0P5KKU9 A0A0P5MYD5 A0A0P4Y9B2 A0A0K8VQ52 A0A0P5EFN5 A0A0P4Y407 A0A0M3QTF2 A0A0P5DWK1
A0A3S2TP60 A0A2A4JBX4 A0A2A4JDE5 E0VQ35 A0A1Y1KFW1 A0A1Y1KC59 A0A2J7RKL3 A0A0K8SN26 A0A146LPP8 A0A0A9ZDF3 A0A1B1M0Q9 A0A2H8TD90 J9K964 A0A224XDW2 A0A182FNT9 A0A1S4FIM7 Q16ZU9 B0WVE0 A0A2S2R1N5 W5JW93 A0A1Q3FPU0 A0A023F3J1 A0A182P4E3 A0A182QRY4 A0A084WSA5 Q7QK54 A0A182UXT1 A0A182HZQ6 A0A182NMX8 A0A182U6A2 A0A182RN68 A0A182XWG9 A0A1J1HLI3 A0A182VYF8 A0A182IR04 A0A182MKF1 A0A182XGM1 A0A0K2TRS2 A0A2L2Y9F6 A0A2P2I1C0 A0A3B0JUD1 A0A1B6L9W4 A0A0P4VVB7 B4GJY3 A0A0P4Z3E1 A0A0N8A8K8 A0A0P5S5Z0 A0A162DFK5 A0A0P5FR53 A0A0P5CM97 A0A0N8CAB5 A0A0N8A3Q2 A0A0P5F7N0 A0A0P5XU95 A0A0N8C4G0 A0A0P5EK42 A0A0P5HG21 A0A0P5AS87 A0A0P4ZTS6 A0A1Y1L6J8 A0A0P4YIM1 A0A0P5UTC7 A0A2C9JV62 A0A0N8CUE6 A0A0P4Y347 A0A0P5EI44 A0A0P5CE47 A0A0P5IWS3 A0A0P5C8R0 A0A0P5VR40 A0A0P5CG86 K1PQ45 A0A0N8A7V3 A0A0P5R6U7 A0A0P5CGB0 A0A0N8BSE3 A0A0P5N2P7 A0A0P5XTT6 A0A0P5K353 A0A0P5CZW4 A0A0K8W363 A0A0P4Y4N3 A0A0P5CS41 A0A0P5WL95 A0A0P4Y4B1 A0A0P6B4D3 A0A0P5KKU9 A0A0P5MYD5 A0A0P4Y9B2 A0A0K8VQ52 A0A0P5EFN5 A0A0P4Y407 A0A0M3QTF2 A0A0P5DWK1
Pubmed
EMBL
ODYU01005430
SOQ46269.1
KQ459053
KPJ04066.1
JTDY01000288
KOB77888.1
+ More
AGBW02011289 OWR46894.1 BABH01018319 BABH01018320 BABH01018321 KQ461073 KPJ09543.1 RSAL01000034 RVE51395.1 NWSH01001952 PCG69575.1 PCG69574.1 DS235389 EEB15491.1 GEZM01086687 JAV59060.1 GEZM01086685 JAV59063.1 NEVH01002717 PNF41376.1 GBRD01011171 JAG54653.1 GDHC01009340 JAQ09289.1 GBHO01001145 JAG42459.1 KT266556 ANS60447.1 GFXV01000272 MBW12077.1 ABLF02024570 GFTR01008448 JAW07978.1 CH477484 EAT40172.1 DS232124 EDS35542.1 GGMS01014640 MBY83843.1 ADMH02000189 ETN67430.1 GFDL01005424 JAV29621.1 GBBI01002826 JAC15886.1 AXCN02001544 ATLV01026419 KE525413 KFB53099.1 AAAB01008799 EAA03707.5 APCN01001998 CVRI01000004 CRK87310.1 AXCM01002302 HACA01011388 CDW28749.1 IAAA01013898 IAAA01013899 LAA04809.1 IACF01002024 LAB67696.1 OUUW01000010 SPP85717.1 GEBQ01019490 JAT20487.1 GDRN01093823 JAI59814.1 CH479184 EDW36949.1 GDIP01219010 JAJ04392.1 GDIP01171873 GDIQ01099340 JAJ51529.1 JAL52386.1 GDIQ01096222 JAL55504.1 LRGB01001581 KZS11044.1 GDIQ01253229 JAJ98495.1 GDIP01184224 JAJ39178.1 GDIP01173914 GDIQ01099339 JAJ49488.1 JAL52387.1 GDIP01185521 JAJ37881.1 GDIP01195875 GDIQ01266809 GDIQ01147863 GDIP01132671 GDIP01108977 GDIQ01052109 JAJ27527.1 JAJ84915.1 GDIP01067478 JAM36237.1 GDIQ01115779 JAL35947.1 GDIP01145320 JAJ78082.1 GDIQ01250530 JAK01195.1 GDIP01198766 JAJ24636.1 GDIP01209762 JAJ13640.1 GEZM01066288 JAV68000.1 GDIP01236366 JAI87035.1 GDIP01108976 JAL94738.1 GDIP01097879 JAM05836.1 GDIP01234471 JAI88930.1 GDIP01148170 GDIQ01052108 JAJ75232.1 JAN42629.1 GDIP01171878 JAJ51524.1 GDIQ01208745 JAK42980.1 GDIP01173912 JAJ49490.1 GDIP01096584 JAM07131.1 GDIP01171916 JAJ51486.1 JH817066 EKC26377.1 GDIP01173913 JAJ49489.1 GDIQ01111868 JAL39858.1 GDIP01171879 JAJ51523.1 GDIQ01149515 JAL02211.1 GDIQ01147862 JAL03864.1 GDIP01067477 JAM36238.1 GDIQ01189897 JAK61828.1 GDIP01179223 JAJ44179.1 GDHF01006805 JAI45509.1 GDIP01236365 JAI87036.1 GDIP01171877 JAJ51525.1 GDIP01084680 JAM19035.1 GDIP01232758 JAI90643.1 GDIP01026696 JAM77019.1 GDIQ01189896 JAK61829.1 GDIQ01149514 JAL02212.1 GDIP01234472 JAI88929.1 GDHF01019158 GDHF01011300 GDHF01000680 JAI33156.1 JAI41014.1 JAI51634.1 GDIP01160874 JAJ62528.1 GDIP01232757 JAI90644.1 CP012523 ALC38779.1 GDIP01154573 JAJ68829.1
AGBW02011289 OWR46894.1 BABH01018319 BABH01018320 BABH01018321 KQ461073 KPJ09543.1 RSAL01000034 RVE51395.1 NWSH01001952 PCG69575.1 PCG69574.1 DS235389 EEB15491.1 GEZM01086687 JAV59060.1 GEZM01086685 JAV59063.1 NEVH01002717 PNF41376.1 GBRD01011171 JAG54653.1 GDHC01009340 JAQ09289.1 GBHO01001145 JAG42459.1 KT266556 ANS60447.1 GFXV01000272 MBW12077.1 ABLF02024570 GFTR01008448 JAW07978.1 CH477484 EAT40172.1 DS232124 EDS35542.1 GGMS01014640 MBY83843.1 ADMH02000189 ETN67430.1 GFDL01005424 JAV29621.1 GBBI01002826 JAC15886.1 AXCN02001544 ATLV01026419 KE525413 KFB53099.1 AAAB01008799 EAA03707.5 APCN01001998 CVRI01000004 CRK87310.1 AXCM01002302 HACA01011388 CDW28749.1 IAAA01013898 IAAA01013899 LAA04809.1 IACF01002024 LAB67696.1 OUUW01000010 SPP85717.1 GEBQ01019490 JAT20487.1 GDRN01093823 JAI59814.1 CH479184 EDW36949.1 GDIP01219010 JAJ04392.1 GDIP01171873 GDIQ01099340 JAJ51529.1 JAL52386.1 GDIQ01096222 JAL55504.1 LRGB01001581 KZS11044.1 GDIQ01253229 JAJ98495.1 GDIP01184224 JAJ39178.1 GDIP01173914 GDIQ01099339 JAJ49488.1 JAL52387.1 GDIP01185521 JAJ37881.1 GDIP01195875 GDIQ01266809 GDIQ01147863 GDIP01132671 GDIP01108977 GDIQ01052109 JAJ27527.1 JAJ84915.1 GDIP01067478 JAM36237.1 GDIQ01115779 JAL35947.1 GDIP01145320 JAJ78082.1 GDIQ01250530 JAK01195.1 GDIP01198766 JAJ24636.1 GDIP01209762 JAJ13640.1 GEZM01066288 JAV68000.1 GDIP01236366 JAI87035.1 GDIP01108976 JAL94738.1 GDIP01097879 JAM05836.1 GDIP01234471 JAI88930.1 GDIP01148170 GDIQ01052108 JAJ75232.1 JAN42629.1 GDIP01171878 JAJ51524.1 GDIQ01208745 JAK42980.1 GDIP01173912 JAJ49490.1 GDIP01096584 JAM07131.1 GDIP01171916 JAJ51486.1 JH817066 EKC26377.1 GDIP01173913 JAJ49489.1 GDIQ01111868 JAL39858.1 GDIP01171879 JAJ51523.1 GDIQ01149515 JAL02211.1 GDIQ01147862 JAL03864.1 GDIP01067477 JAM36238.1 GDIQ01189897 JAK61828.1 GDIP01179223 JAJ44179.1 GDHF01006805 JAI45509.1 GDIP01236365 JAI87036.1 GDIP01171877 JAJ51525.1 GDIP01084680 JAM19035.1 GDIP01232758 JAI90643.1 GDIP01026696 JAM77019.1 GDIQ01189896 JAK61829.1 GDIQ01149514 JAL02212.1 GDIP01234472 JAI88929.1 GDHF01019158 GDHF01011300 GDHF01000680 JAI33156.1 JAI41014.1 JAI51634.1 GDIP01160874 JAJ62528.1 GDIP01232757 JAI90644.1 CP012523 ALC38779.1 GDIP01154573 JAJ68829.1
Proteomes
UP000053268
UP000037510
UP000007151
UP000005204
UP000053240
UP000283053
+ More
UP000218220 UP000009046 UP000235965 UP000007819 UP000069272 UP000008820 UP000002320 UP000000673 UP000075885 UP000075886 UP000030765 UP000007062 UP000075903 UP000075840 UP000075884 UP000075902 UP000075900 UP000076408 UP000183832 UP000075920 UP000075880 UP000075883 UP000076407 UP000268350 UP000008744 UP000076858 UP000076420 UP000005408 UP000092553
UP000218220 UP000009046 UP000235965 UP000007819 UP000069272 UP000008820 UP000002320 UP000000673 UP000075885 UP000075886 UP000030765 UP000007062 UP000075903 UP000075840 UP000075884 UP000075902 UP000075900 UP000076408 UP000183832 UP000075920 UP000075880 UP000075883 UP000076407 UP000268350 UP000008744 UP000076858 UP000076420 UP000005408 UP000092553
PRIDE
Interpro
IPR001881
EGF-like_Ca-bd_dom
+ More
IPR008859 Thrombospondin_C
IPR013320 ConA-like_dom_sf
IPR017897 Thrombospondin_3_rpt
IPR009030 Growth_fac_rcpt_cys_sf
IPR018097 EGF_Ca-bd_CS
IPR003367 Thrombospondin_3-like_rpt
IPR000742 EGF-like_dom
IPR028974 TSP_type-3_rpt
IPR013032 EGF-like_CS
IPR024665 Thbs/COMP_coiled-coil
IPR028507 Thrombospondin-3
IPR026823 cEGF
IPR000867 IGFBP-like
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR008859 Thrombospondin_C
IPR013320 ConA-like_dom_sf
IPR017897 Thrombospondin_3_rpt
IPR009030 Growth_fac_rcpt_cys_sf
IPR018097 EGF_Ca-bd_CS
IPR003367 Thrombospondin_3-like_rpt
IPR000742 EGF-like_dom
IPR028974 TSP_type-3_rpt
IPR013032 EGF-like_CS
IPR024665 Thbs/COMP_coiled-coil
IPR028507 Thrombospondin-3
IPR026823 cEGF
IPR000867 IGFBP-like
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
Gene 3D
ProteinModelPortal
A0A2H1VZK9
A0A194QKU2
A0A0L7LQW3
A0A212EZM8
H9IX53
A0A194QVH8
+ More
A0A3S2TP60 A0A2A4JBX4 A0A2A4JDE5 E0VQ35 A0A1Y1KFW1 A0A1Y1KC59 A0A2J7RKL3 A0A0K8SN26 A0A146LPP8 A0A0A9ZDF3 A0A1B1M0Q9 A0A2H8TD90 J9K964 A0A224XDW2 A0A182FNT9 A0A1S4FIM7 Q16ZU9 B0WVE0 A0A2S2R1N5 W5JW93 A0A1Q3FPU0 A0A023F3J1 A0A182P4E3 A0A182QRY4 A0A084WSA5 Q7QK54 A0A182UXT1 A0A182HZQ6 A0A182NMX8 A0A182U6A2 A0A182RN68 A0A182XWG9 A0A1J1HLI3 A0A182VYF8 A0A182IR04 A0A182MKF1 A0A182XGM1 A0A0K2TRS2 A0A2L2Y9F6 A0A2P2I1C0 A0A3B0JUD1 A0A1B6L9W4 A0A0P4VVB7 B4GJY3 A0A0P4Z3E1 A0A0N8A8K8 A0A0P5S5Z0 A0A162DFK5 A0A0P5FR53 A0A0P5CM97 A0A0N8CAB5 A0A0N8A3Q2 A0A0P5F7N0 A0A0P5XU95 A0A0N8C4G0 A0A0P5EK42 A0A0P5HG21 A0A0P5AS87 A0A0P4ZTS6 A0A1Y1L6J8 A0A0P4YIM1 A0A0P5UTC7 A0A2C9JV62 A0A0N8CUE6 A0A0P4Y347 A0A0P5EI44 A0A0P5CE47 A0A0P5IWS3 A0A0P5C8R0 A0A0P5VR40 A0A0P5CG86 K1PQ45 A0A0N8A7V3 A0A0P5R6U7 A0A0P5CGB0 A0A0N8BSE3 A0A0P5N2P7 A0A0P5XTT6 A0A0P5K353 A0A0P5CZW4 A0A0K8W363 A0A0P4Y4N3 A0A0P5CS41 A0A0P5WL95 A0A0P4Y4B1 A0A0P6B4D3 A0A0P5KKU9 A0A0P5MYD5 A0A0P4Y9B2 A0A0K8VQ52 A0A0P5EFN5 A0A0P4Y407 A0A0M3QTF2 A0A0P5DWK1
A0A3S2TP60 A0A2A4JBX4 A0A2A4JDE5 E0VQ35 A0A1Y1KFW1 A0A1Y1KC59 A0A2J7RKL3 A0A0K8SN26 A0A146LPP8 A0A0A9ZDF3 A0A1B1M0Q9 A0A2H8TD90 J9K964 A0A224XDW2 A0A182FNT9 A0A1S4FIM7 Q16ZU9 B0WVE0 A0A2S2R1N5 W5JW93 A0A1Q3FPU0 A0A023F3J1 A0A182P4E3 A0A182QRY4 A0A084WSA5 Q7QK54 A0A182UXT1 A0A182HZQ6 A0A182NMX8 A0A182U6A2 A0A182RN68 A0A182XWG9 A0A1J1HLI3 A0A182VYF8 A0A182IR04 A0A182MKF1 A0A182XGM1 A0A0K2TRS2 A0A2L2Y9F6 A0A2P2I1C0 A0A3B0JUD1 A0A1B6L9W4 A0A0P4VVB7 B4GJY3 A0A0P4Z3E1 A0A0N8A8K8 A0A0P5S5Z0 A0A162DFK5 A0A0P5FR53 A0A0P5CM97 A0A0N8CAB5 A0A0N8A3Q2 A0A0P5F7N0 A0A0P5XU95 A0A0N8C4G0 A0A0P5EK42 A0A0P5HG21 A0A0P5AS87 A0A0P4ZTS6 A0A1Y1L6J8 A0A0P4YIM1 A0A0P5UTC7 A0A2C9JV62 A0A0N8CUE6 A0A0P4Y347 A0A0P5EI44 A0A0P5CE47 A0A0P5IWS3 A0A0P5C8R0 A0A0P5VR40 A0A0P5CG86 K1PQ45 A0A0N8A7V3 A0A0P5R6U7 A0A0P5CGB0 A0A0N8BSE3 A0A0P5N2P7 A0A0P5XTT6 A0A0P5K353 A0A0P5CZW4 A0A0K8W363 A0A0P4Y4N3 A0A0P5CS41 A0A0P5WL95 A0A0P4Y4B1 A0A0P6B4D3 A0A0P5KKU9 A0A0P5MYD5 A0A0P4Y9B2 A0A0K8VQ52 A0A0P5EFN5 A0A0P4Y407 A0A0M3QTF2 A0A0P5DWK1
PDB
3FBY
E-value=1.64737e-134,
Score=1232
Ontologies
PATHWAY
GO
PANTHER
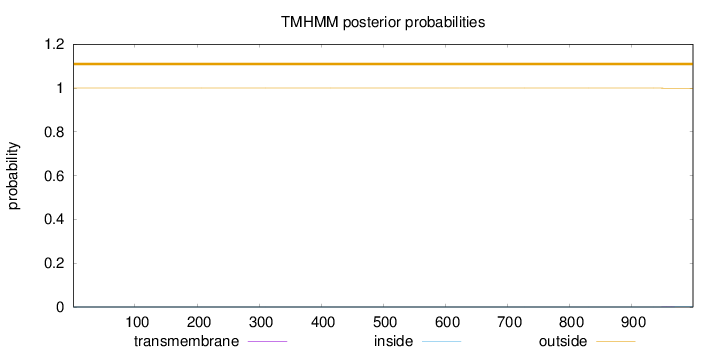
Topology
Length:
999
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01663
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00001
outside
1 - 999
Population Genetic Test Statistics
Pi
170.943703
Theta
158.157068
Tajima's D
0.537875
CLR
0.239226
CSRT
0.526523673816309
Interpretation
Uncertain
