Gene
KWMTBOMO11292
Annotation
PREDICTED:_cyclin-dependent_kinases_regulatory_subunit_[Amyelois_transitella]
Full name
Cyclin-dependent kinases regulatory subunit
Alternative Name
SUC1 homolog
Location in the cell
Extracellular Reliability : 1.212 Nuclear Reliability : 1.715
Sequence
CDS
ATGTCGAGAGACATTTATTATTCCCAAAAATATTATGACGAAGAACATGAATACAGACATGTAGTACTACCAAAAGAGATGGTAAAGTTAGTGCCAAAAAACCATCTAATGTCAGAACAAGAGTGGCGCAGCATCGGTGTGCAGCAAAGTCAAGGTTGGGTGCACTATATGACACATCAGCCAGAACCACATATTTTATTATTCCGAAGGAGGATAACAACACCACCAGAAAAAAATTGA
Protein
MSRDIYYSQKYYDEEHEYRHVVLPKEMVKLVPKNHLMSEQEWRSIGVQQSQGWVHYMTHQPEPHILLFRRRITTPPEKN
Summary
Description
Binds to the catalytic subunit of the cyclin dependent kinases and is essential for their biological function.
Subunit
Forms a homohexamer that can probably bind six kinase subunits.
Similarity
Belongs to the CKS family.
Keywords
Cell cycle
Cell division
Complete proteome
Reference proteome
Feature
chain Cyclin-dependent kinases regulatory subunit
Uniprot
A0A212EZL5
A0A3S2M4T1
A0A194QW71
A0A0U1SG36
A0A2A4JCL6
A0A2H1WKS4
+ More
S4PWW7 A0A139WER6 A0A0J7K7M5 A0A0M9AB01 A0A067QX59 A0A195AWN3 A0A158NHV6 A0A088A623 A0A0L0BKX6 A0A3L8D7B1 A0A1B0B7J2 A0A1B0FGE2 A0A336KDX9 C3YNN9 A0A1P8D432 B3MUT3 A0A2P8XU45 A0A1I8N9K6 A0A1I8NQQ5 A0A182NEL1 A0A182VUT4 B4M9T1 A0A0M3QTP6 B4JPW8 B5DH81 T1DQP5 W5J4B5 A0A182F5A7 A0A1B2AK90 B4NY24 B4HYW3 B3N7Y6 B4Q7R0 Q24152 B4MVI0 A0A182XIN9 Q7PTN0 A0A182HG48 A0A162CIV1 A0A182RGA0 B4KEZ7 C0KQV9 A0A023EDZ1 A0A0P6CT35 Q0IFN1 T1H4R9 A0A1Q3EVU4 V3ZY62 B0W239 A0A182PJ09 A0A1W4VI04 P41384 A0A182H5Y0 N6URP3 C1BSQ6 E0VQ27 Q962Y1 A0A131XB52 A0A3S3QD61 T1IFE5 A0A1W4WR78 A0A1B0GMH2 A0A0C9RZZ0 A0A023G5K3 G3MQ74 A0A224YV38 A0A131YEA4 L7M5K5 A0A224Y5K9 C1BZV7 A0A0B6Y974 F6WFW0 B7Q8G6 A0A023FY58 A0A3S3SER3 A0A226MGL3 A0A218UMY1 A0A226MG49 A0A1L1S0Y9 A0A069DMD5 A0A0K8R643 A0A1L8DWW6 A0A1J1I3J5 A0A3B5KJ13 A0A3M7RAZ8 T1L0L7 A0A0K8TSI7 A0A087U8Q0 U3JEW3 A0A194QGH3 U3EB95 A0A1E1WZB1 A0A3M0J286 B5FXN4
S4PWW7 A0A139WER6 A0A0J7K7M5 A0A0M9AB01 A0A067QX59 A0A195AWN3 A0A158NHV6 A0A088A623 A0A0L0BKX6 A0A3L8D7B1 A0A1B0B7J2 A0A1B0FGE2 A0A336KDX9 C3YNN9 A0A1P8D432 B3MUT3 A0A2P8XU45 A0A1I8N9K6 A0A1I8NQQ5 A0A182NEL1 A0A182VUT4 B4M9T1 A0A0M3QTP6 B4JPW8 B5DH81 T1DQP5 W5J4B5 A0A182F5A7 A0A1B2AK90 B4NY24 B4HYW3 B3N7Y6 B4Q7R0 Q24152 B4MVI0 A0A182XIN9 Q7PTN0 A0A182HG48 A0A162CIV1 A0A182RGA0 B4KEZ7 C0KQV9 A0A023EDZ1 A0A0P6CT35 Q0IFN1 T1H4R9 A0A1Q3EVU4 V3ZY62 B0W239 A0A182PJ09 A0A1W4VI04 P41384 A0A182H5Y0 N6URP3 C1BSQ6 E0VQ27 Q962Y1 A0A131XB52 A0A3S3QD61 T1IFE5 A0A1W4WR78 A0A1B0GMH2 A0A0C9RZZ0 A0A023G5K3 G3MQ74 A0A224YV38 A0A131YEA4 L7M5K5 A0A224Y5K9 C1BZV7 A0A0B6Y974 F6WFW0 B7Q8G6 A0A023FY58 A0A3S3SER3 A0A226MGL3 A0A218UMY1 A0A226MG49 A0A1L1S0Y9 A0A069DMD5 A0A0K8R643 A0A1L8DWW6 A0A1J1I3J5 A0A3B5KJ13 A0A3M7RAZ8 T1L0L7 A0A0K8TSI7 A0A087U8Q0 U3JEW3 A0A194QGH3 U3EB95 A0A1E1WZB1 A0A3M0J286 B5FXN4
Pubmed
22118469
26354079
23622113
18362917
19820115
24845553
+ More
21347285 26108605 30249741 18563158 17994087 29403074 25315136 15632085 20920257 23761445 17550304 22936249 7809159 10731132 12537572 12537569 12364791 14747013 17210077 24945155 17510324 23254933 8180002 26483478 23537049 20566863 28049606 26131772 22216098 28797301 26830274 25576852 12481130 15114417 24621616 15592404 26334808 21551351 30375419 26369729 25243066 28503490 17018643
21347285 26108605 30249741 18563158 17994087 29403074 25315136 15632085 20920257 23761445 17550304 22936249 7809159 10731132 12537572 12537569 12364791 14747013 17210077 24945155 17510324 23254933 8180002 26483478 23537049 20566863 28049606 26131772 22216098 28797301 26830274 25576852 12481130 15114417 24621616 15592404 26334808 21551351 30375419 26369729 25243066 28503490 17018643
EMBL
AGBW02011289
OWR46884.1
RSAL01000034
RVE51403.1
KQ461073
KPJ09554.1
+ More
JQ287498 AFO67941.1 NWSH01001952 PCG69589.1 ODYU01009288 SOQ53607.1 GAIX01006068 JAA86492.1 KQ971354 KYB26351.1 LBMM01012503 KMQ86191.1 KQ435697 KOX80790.1 KK853253 KDR09304.1 KQ976725 KYM76581.1 ADTU01016073 ADTU01016074 JRES01001704 JRES01000864 KNC20750.1 KNC27687.1 QOIP01000012 RLU15768.1 JXJN01009677 CCAG010000709 UFQS01000346 UFQT01000346 SSX03066.1 SSX23432.1 GG666535 EEN57999.1 KX015775 APM86330.1 CH902624 EDV32998.1 PYGN01001345 PSN35519.1 CH940654 EDW57957.1 CP012523 ALC39249.1 CH916372 EDV98948.1 CH379058 EDY69656.1 GAMD01002344 JAA99246.1 ADMH02002093 GGFL01004723 ETN59287.1 MBW68901.1 KX531758 ANY27568.1 CM000157 EDW88626.1 CH480818 EDW52243.1 CH954177 EDV58347.1 CM000361 CM002910 EDX04342.1 KMY89240.1 U40077 AE014134 AY061285 CH963857 EDW75700.2 AAAB01008797 EAA03636.4 APCN01004115 LRGB01000725 KZS16322.1 CH933807 EDW11966.1 FJ623275 ACM69341.1 GAPW01006764 JAC06834.1 GDIP01009554 JAM94161.1 CH477306 EAT44112.1 CAQQ02032992 GFDL01015616 JAV19429.1 KB202883 ESO87580.1 DS231825 DS232411 EDS28167.1 EDS41290.1 Z28352 JXUM01112929 KQ565638 KXJ70825.1 APGK01019290 KB740098 KB631733 ENN81407.1 ERL85651.1 BT077635 BT121504 HACA01002502 ACO12059.1 ADD38434.1 CDW19863.1 DS235389 EEB15483.1 AF395863 AAK91295.2 GEFH01004949 JAP63632.1 NCKU01000025 RWS17871.1 ACPB03001633 AJVK01024542 GBZX01002891 JAG89849.1 GBBM01006376 JAC29042.1 JO844025 AEO35642.1 GFPF01007017 MAA18163.1 GEDV01011679 JAP76878.1 GACK01006591 JAA58443.1 GFTR01000647 JAW15779.1 BT080136 ACO14560.1 HACG01005105 CEK51970.1 ABJB010229219 ABJB011032816 DS883946 EEC15138.1 GBBL01000906 JAC26414.1 NCKU01001080 NCKU01001077 NCKU01000937 RWS13147.1 RWS13163.1 RWS13605.1 MCFN01000972 OXB54169.1 MUZQ01000219 OWK55095.1 AWGT02008889 OXB54255.1 AADN05000763 GBGD01003671 JAC85218.1 GADI01007455 JAA66353.1 GFDF01003156 JAV10928.1 CVRI01000038 CRK94338.1 REGN01003816 RNA20574.1 CAEY01000867 GDAI01000698 JAI16905.1 KK118740 KFM73739.1 AGTO01012145 KQ459053 KPJ04055.1 GAMP01004562 JAB48193.1 GFAC01006833 JAT92355.1 QRBI01000191 RMB94988.1 DQ213506 DQ213507 ACH43795.1
JQ287498 AFO67941.1 NWSH01001952 PCG69589.1 ODYU01009288 SOQ53607.1 GAIX01006068 JAA86492.1 KQ971354 KYB26351.1 LBMM01012503 KMQ86191.1 KQ435697 KOX80790.1 KK853253 KDR09304.1 KQ976725 KYM76581.1 ADTU01016073 ADTU01016074 JRES01001704 JRES01000864 KNC20750.1 KNC27687.1 QOIP01000012 RLU15768.1 JXJN01009677 CCAG010000709 UFQS01000346 UFQT01000346 SSX03066.1 SSX23432.1 GG666535 EEN57999.1 KX015775 APM86330.1 CH902624 EDV32998.1 PYGN01001345 PSN35519.1 CH940654 EDW57957.1 CP012523 ALC39249.1 CH916372 EDV98948.1 CH379058 EDY69656.1 GAMD01002344 JAA99246.1 ADMH02002093 GGFL01004723 ETN59287.1 MBW68901.1 KX531758 ANY27568.1 CM000157 EDW88626.1 CH480818 EDW52243.1 CH954177 EDV58347.1 CM000361 CM002910 EDX04342.1 KMY89240.1 U40077 AE014134 AY061285 CH963857 EDW75700.2 AAAB01008797 EAA03636.4 APCN01004115 LRGB01000725 KZS16322.1 CH933807 EDW11966.1 FJ623275 ACM69341.1 GAPW01006764 JAC06834.1 GDIP01009554 JAM94161.1 CH477306 EAT44112.1 CAQQ02032992 GFDL01015616 JAV19429.1 KB202883 ESO87580.1 DS231825 DS232411 EDS28167.1 EDS41290.1 Z28352 JXUM01112929 KQ565638 KXJ70825.1 APGK01019290 KB740098 KB631733 ENN81407.1 ERL85651.1 BT077635 BT121504 HACA01002502 ACO12059.1 ADD38434.1 CDW19863.1 DS235389 EEB15483.1 AF395863 AAK91295.2 GEFH01004949 JAP63632.1 NCKU01000025 RWS17871.1 ACPB03001633 AJVK01024542 GBZX01002891 JAG89849.1 GBBM01006376 JAC29042.1 JO844025 AEO35642.1 GFPF01007017 MAA18163.1 GEDV01011679 JAP76878.1 GACK01006591 JAA58443.1 GFTR01000647 JAW15779.1 BT080136 ACO14560.1 HACG01005105 CEK51970.1 ABJB010229219 ABJB011032816 DS883946 EEC15138.1 GBBL01000906 JAC26414.1 NCKU01001080 NCKU01001077 NCKU01000937 RWS13147.1 RWS13163.1 RWS13605.1 MCFN01000972 OXB54169.1 MUZQ01000219 OWK55095.1 AWGT02008889 OXB54255.1 AADN05000763 GBGD01003671 JAC85218.1 GADI01007455 JAA66353.1 GFDF01003156 JAV10928.1 CVRI01000038 CRK94338.1 REGN01003816 RNA20574.1 CAEY01000867 GDAI01000698 JAI16905.1 KK118740 KFM73739.1 AGTO01012145 KQ459053 KPJ04055.1 GAMP01004562 JAB48193.1 GFAC01006833 JAT92355.1 QRBI01000191 RMB94988.1 DQ213506 DQ213507 ACH43795.1
Proteomes
UP000007151
UP000283053
UP000053240
UP000218220
UP000007266
UP000036403
+ More
UP000053105 UP000027135 UP000078540 UP000005205 UP000005203 UP000037069 UP000279307 UP000092460 UP000092444 UP000001554 UP000007801 UP000245037 UP000095301 UP000095300 UP000075884 UP000075920 UP000008792 UP000092553 UP000001070 UP000001819 UP000000673 UP000069272 UP000002282 UP000001292 UP000008711 UP000000304 UP000000803 UP000007798 UP000076407 UP000007062 UP000075840 UP000076858 UP000075900 UP000009192 UP000008820 UP000015102 UP000030746 UP000002320 UP000075885 UP000192221 UP000069940 UP000249989 UP000019118 UP000030742 UP000009046 UP000285301 UP000015103 UP000192223 UP000092462 UP000008144 UP000001555 UP000198323 UP000197619 UP000198419 UP000000539 UP000183832 UP000005226 UP000276133 UP000015104 UP000054359 UP000016665 UP000053268 UP000269221
UP000053105 UP000027135 UP000078540 UP000005205 UP000005203 UP000037069 UP000279307 UP000092460 UP000092444 UP000001554 UP000007801 UP000245037 UP000095301 UP000095300 UP000075884 UP000075920 UP000008792 UP000092553 UP000001070 UP000001819 UP000000673 UP000069272 UP000002282 UP000001292 UP000008711 UP000000304 UP000000803 UP000007798 UP000076407 UP000007062 UP000075840 UP000076858 UP000075900 UP000009192 UP000008820 UP000015102 UP000030746 UP000002320 UP000075885 UP000192221 UP000069940 UP000249989 UP000019118 UP000030742 UP000009046 UP000285301 UP000015103 UP000192223 UP000092462 UP000008144 UP000001555 UP000198323 UP000197619 UP000198419 UP000000539 UP000183832 UP000005226 UP000276133 UP000015104 UP000054359 UP000016665 UP000053268 UP000269221
PRIDE
Pfam
PF01111 CKS
SUPFAM
SSF55637
SSF55637
Gene 3D
ProteinModelPortal
A0A212EZL5
A0A3S2M4T1
A0A194QW71
A0A0U1SG36
A0A2A4JCL6
A0A2H1WKS4
+ More
S4PWW7 A0A139WER6 A0A0J7K7M5 A0A0M9AB01 A0A067QX59 A0A195AWN3 A0A158NHV6 A0A088A623 A0A0L0BKX6 A0A3L8D7B1 A0A1B0B7J2 A0A1B0FGE2 A0A336KDX9 C3YNN9 A0A1P8D432 B3MUT3 A0A2P8XU45 A0A1I8N9K6 A0A1I8NQQ5 A0A182NEL1 A0A182VUT4 B4M9T1 A0A0M3QTP6 B4JPW8 B5DH81 T1DQP5 W5J4B5 A0A182F5A7 A0A1B2AK90 B4NY24 B4HYW3 B3N7Y6 B4Q7R0 Q24152 B4MVI0 A0A182XIN9 Q7PTN0 A0A182HG48 A0A162CIV1 A0A182RGA0 B4KEZ7 C0KQV9 A0A023EDZ1 A0A0P6CT35 Q0IFN1 T1H4R9 A0A1Q3EVU4 V3ZY62 B0W239 A0A182PJ09 A0A1W4VI04 P41384 A0A182H5Y0 N6URP3 C1BSQ6 E0VQ27 Q962Y1 A0A131XB52 A0A3S3QD61 T1IFE5 A0A1W4WR78 A0A1B0GMH2 A0A0C9RZZ0 A0A023G5K3 G3MQ74 A0A224YV38 A0A131YEA4 L7M5K5 A0A224Y5K9 C1BZV7 A0A0B6Y974 F6WFW0 B7Q8G6 A0A023FY58 A0A3S3SER3 A0A226MGL3 A0A218UMY1 A0A226MG49 A0A1L1S0Y9 A0A069DMD5 A0A0K8R643 A0A1L8DWW6 A0A1J1I3J5 A0A3B5KJ13 A0A3M7RAZ8 T1L0L7 A0A0K8TSI7 A0A087U8Q0 U3JEW3 A0A194QGH3 U3EB95 A0A1E1WZB1 A0A3M0J286 B5FXN4
S4PWW7 A0A139WER6 A0A0J7K7M5 A0A0M9AB01 A0A067QX59 A0A195AWN3 A0A158NHV6 A0A088A623 A0A0L0BKX6 A0A3L8D7B1 A0A1B0B7J2 A0A1B0FGE2 A0A336KDX9 C3YNN9 A0A1P8D432 B3MUT3 A0A2P8XU45 A0A1I8N9K6 A0A1I8NQQ5 A0A182NEL1 A0A182VUT4 B4M9T1 A0A0M3QTP6 B4JPW8 B5DH81 T1DQP5 W5J4B5 A0A182F5A7 A0A1B2AK90 B4NY24 B4HYW3 B3N7Y6 B4Q7R0 Q24152 B4MVI0 A0A182XIN9 Q7PTN0 A0A182HG48 A0A162CIV1 A0A182RGA0 B4KEZ7 C0KQV9 A0A023EDZ1 A0A0P6CT35 Q0IFN1 T1H4R9 A0A1Q3EVU4 V3ZY62 B0W239 A0A182PJ09 A0A1W4VI04 P41384 A0A182H5Y0 N6URP3 C1BSQ6 E0VQ27 Q962Y1 A0A131XB52 A0A3S3QD61 T1IFE5 A0A1W4WR78 A0A1B0GMH2 A0A0C9RZZ0 A0A023G5K3 G3MQ74 A0A224YV38 A0A131YEA4 L7M5K5 A0A224Y5K9 C1BZV7 A0A0B6Y974 F6WFW0 B7Q8G6 A0A023FY58 A0A3S3SER3 A0A226MGL3 A0A218UMY1 A0A226MG49 A0A1L1S0Y9 A0A069DMD5 A0A0K8R643 A0A1L8DWW6 A0A1J1I3J5 A0A3B5KJ13 A0A3M7RAZ8 T1L0L7 A0A0K8TSI7 A0A087U8Q0 U3JEW3 A0A194QGH3 U3EB95 A0A1E1WZB1 A0A3M0J286 B5FXN4
PDB
1DKT
E-value=1.55904e-21,
Score=247
Ontologies
GO
GO:0016301
GO:0016538
GO:0051301
GO:0007049
GO:0019005
GO:0061575
GO:0042393
GO:0000307
GO:0019901
GO:0043130
GO:0007346
GO:0007488
GO:0007057
GO:0006468
GO:0031145
GO:0035186
GO:0016740
GO:0007144
GO:0007143
GO:0051225
GO:0016021
GO:0006270
GO:0042555
GO:0006260
GO:0006811
GO:0005622
GO:0006810
GO:0003824
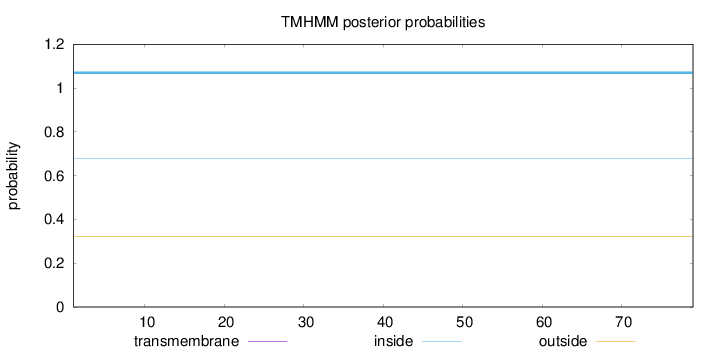
Topology
Length:
79
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.67792
inside
1 - 79
Population Genetic Test Statistics
Pi
178.65849
Theta
176.971853
Tajima's D
0.094636
CLR
0.003999
CSRT
0.395080245987701
Interpretation
Uncertain
