Gene
KWMTBOMO11281 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002755
Annotation
SUMO-1_activating_enzyme_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.286
Sequence
CDS
ATGGTTGGAAATAATGAAGTCGAACTTTCTGAGGCAGAAGCCGAACAGTACGACCGACAAATCCGCCTGTGGGGCTTAGACTCGCAGAAGAGACTACGTGCAGCGAAAGTATTAATCATCGGCTTGTCCGGACTCGGCGCTGAAATTGCAAAAAACGTAATCCTCACCGGTGTAAAAAGTGTGTGTTTGTTAGACAATGAGAAACTGAAACAAATCGATCTGTACTCCCAATTTCTGTGTCCTCCTGACAAGATCGGCGTAAATCGAGCAGAGGGTTCGTTGGAAAGAGCTCGAGGCTTGAATCCGATGGTCGATGTCACAAGTCATACTAAGGGCGTAGATGAGCTACCTGACAGCTTCTTCACGGAATTTGACGTCGTATGTGCAACAGGTCTCAAACAAGAGCAATTCGAACGAATCAACAATGCTTGTCGCGACAGCAACAAAAAGTTTATCTGCGGCGACGTCTGGGGTACGTACGGTTACATGTTCTCTGATCTAGTTGATCACGAGTACTCCGAGGAGATTGTACAACACAAAGCCACCAAACGTGGGCCCGATGATGAAGAAAAAAATGCCCGAGAAACAGTTTCTATAACAGTAAAACGTCGAGCTATATATGTCCCGCTTCAGAATGCTTTATCAGCTGATTGGAATTCACCAGAGATGCGTTCTAGGCTACGTCGTGGTGATTGTGGCTATTTTGCTATGAAGCTCCTGTTACGCTTCAGAGATGAATATAACAGGAATCCTGACCCGGCTCGAAGAAAAGACGACATTAATTTACTACTGAAAATGCGTGATGAAATAGTTCGTGAATTGTCACTTCCAGCGAGTTTCATCAAAGATTCGTTGTTTTATGATGTGTTTGGAGTTGTGGCAGCAGCTGCTGCTGTTATCGGTGGTGTTATTGGCCAGGAGGTGGTCAAAGCTGTCAGTCAGAGGGAACCACCACATAACAATATGTTCTTCTTCAATCCAATTAAATGCTTAGGTTATACTGAACTGTATGGTCAATGA
Protein
MVGNNEVELSEAEAEQYDRQIRLWGLDSQKRLRAAKVLIIGLSGLGAEIAKNVILTGVKSVCLLDNEKLKQIDLYSQFLCPPDKIGVNRAEGSLERARGLNPMVDVTSHTKGVDELPDSFFTEFDVVCATGLKQEQFERINNACRDSNKKFICGDVWGTYGYMFSDLVDHEYSEEIVQHKATKRGPDDEEKNARETVSITVKRRAIYVPLQNALSADWNSPEMRSRLRRGDCGYFAMKLLLRFRDEYNRNPDPARRKDDINLLLKMRDEIVRELSLPASFIKDSLFYDVFGVVAAAAAVIGGVIGQEVVKAVSQREPPHNNMFFFNPIKCLGYTELYGQ
Summary
Uniprot
Q1HPK7
A0A0L7LCH9
A0A2A4K0K2
A0A2H1WYD2
A0A194QEZ3
A0A2Z5U813
+ More
A0A194QVR4 S4P5E5 A0A212FNP5 D8VH47 D8VH16 D8VH30 D8VGZ0 D8VH14 D8VGZ2 D8VGZ9 D8VH10 G9JR83 G9JR82 D8VGZ1 G9JR97 G9JRA5 G9JR92 G9JR67 G9JR68 G9JQ18 D8VH31 G9JQ39 G9JQ66 G9JRA2 G9JR71 G9JR76 G9JRA7 G9JRB0 G9JRC0 G9JR80 G9JR62 G9JRB3 G9JQ28 G9JQ57 G9JQ03 G9JQ47 G9JR84 G9JQ51 G9JQ49 G9JQ30 G9JQ24 G9JQ36 G9JQ68 G9JQ43 G9JQ17 G9JQ16 G9JQ12 G9JQ40 G9JQ65 G9JRA1 G9JQ46 G9JQ19 G9JQ08 G9JQ37 G9JQ38 G9JR85 D8VH19 G9JQ35 G9JQ33 G9JPZ2 G9JR79 G9JR88 G9JR94 G9JR63 G9JRB4 D8VH22 D8VH40 D8VH06 D8VH04 G9JR70 G9JRB7 G9JR74 G9JRB1 G9JR69 G9JR75 D8VH08 G9JRB5 G9JRA9 D8VH12 G9JRC1 G9JQ50 G9JR91 G9JQ10 G9JQ05 G9JQ34 G9JQ54 G9JQ58 G9JQ25 G9JQ64 G9JPZ3 G9JQ67 G9JQ44 G9JQ23 G9JQ20
A0A194QVR4 S4P5E5 A0A212FNP5 D8VH47 D8VH16 D8VH30 D8VGZ0 D8VH14 D8VGZ2 D8VGZ9 D8VH10 G9JR83 G9JR82 D8VGZ1 G9JR97 G9JRA5 G9JR92 G9JR67 G9JR68 G9JQ18 D8VH31 G9JQ39 G9JQ66 G9JRA2 G9JR71 G9JR76 G9JRA7 G9JRB0 G9JRC0 G9JR80 G9JR62 G9JRB3 G9JQ28 G9JQ57 G9JQ03 G9JQ47 G9JR84 G9JQ51 G9JQ49 G9JQ30 G9JQ24 G9JQ36 G9JQ68 G9JQ43 G9JQ17 G9JQ16 G9JQ12 G9JQ40 G9JQ65 G9JRA1 G9JQ46 G9JQ19 G9JQ08 G9JQ37 G9JQ38 G9JR85 D8VH19 G9JQ35 G9JQ33 G9JPZ2 G9JR79 G9JR88 G9JR94 G9JR63 G9JRB4 D8VH22 D8VH40 D8VH06 D8VH04 G9JR70 G9JRB7 G9JR74 G9JRB1 G9JR69 G9JR75 D8VH08 G9JRB5 G9JRA9 D8VH12 G9JRC1 G9JQ50 G9JR91 G9JQ10 G9JQ05 G9JQ34 G9JQ54 G9JQ58 G9JQ25 G9JQ64 G9JPZ3 G9JQ67 G9JQ44 G9JQ23 G9JQ20
EMBL
BABH01039963
DQ443395
ABF51484.1
JTDY01001778
KOB72901.1
NWSH01000348
+ More
PCG77172.1 ODYU01012011 SOQ58079.1 KQ459053 KPJ04047.1 AP017484 BBB06740.1 KQ461073 KPJ09562.1 GAIX01005594 JAA86966.1 AGBW02004849 OWR55364.1 GQ986284 ADI82471.1 GQ986253 GQ986263 ADI82440.1 GQ986267 GQ986269 GQ986270 GQ986271 GQ986273 GQ986274 GQ986275 GQ986281 GQ986283 GQ986285 GQ986286 ADI82454.1 GQ986227 GQ986230 GQ986231 GQ986232 GQ986233 GQ986235 GQ986237 GQ986238 GQ986239 GQ986240 ADI82414.1 GQ986251 GQ986255 ADI82438.1 GQ986229 GQ986234 ADI82416.1 GQ986236 ADI82423.1 GQ986247 GQ986248 GQ986252 GQ986254 GQ986257 GQ986258 GQ986261 GQ986262 GQ986264 ADI82434.1 JN898214 AET96867.1 JN898213 AET96866.1 GQ986228 ADI82415.1 JN898228 AET96881.1 JN898236 AET96889.1 JN898223 AET96876.1 JN898198 JN898224 AET96851.1 JN898199 JN898208 AET96852.1 JN897799 AET96452.1 GQ986268 GQ986272 GQ986276 GQ986282 ADI82455.1 JN897820 AET96473.1 JN897847 AET96500.1 JN898233 AET96886.1 JN898202 AET96855.1 JN898207 JN898250 AET96860.1 JN898235 JN898238 AET96891.1 JN898239 JN898241 JN898243 AET96894.1 JN898251 AET96904.1 JN898211 JN898249 AET96864.1 JN898192 JN898193 AET96846.1 JN898244 JN898247 AET96897.1 JN897809 AET96462.1 JN897838 AET96491.1 JN897784 AET96437.1 JN897828 AET96481.1 JN898215 AET96868.1 JN897832 AET96485.1 JN897830 AET96483.1 JN897811 AET96464.1 JN897805 AET96458.1 JN897817 AET96470.1 JN897849 AET96502.1 JN897823 JN897824 AET96477.1 JN897798 JN897802 AET96451.1 JN897783 JN897795 JN897796 JN897797 AET96450.1 JN897785 JN897787 JN897788 JN897792 JN897793 JN897794 JN897810 JN897840 JN897842 AET96446.1 JN897821 JN897822 AET96474.1 JN897775 JN897846 AET96499.1 JN898232 AET96885.1 JN897827 AET96480.1 JN897800 AET96453.1 JN897789 AET96442.1 JN897818 AET96471.1 JN897819 AET96472.1 JN898216 AET96869.1 GQ986256 GQ986265 GQ986266 ADI82443.1 JN897816 AET96469.1 JN897814 AET96467.1 JN897773 AET96426.1 JN898210 AET96863.1 JN898217 JN898218 JN898219 JN898220 JN898221 AET96872.1 JN898225 JN898226 JN898227 AET96878.1 JN898191 JN898194 AET96847.1 JN898245 AET96898.1 GQ986259 ADI82446.1 GQ986277 GQ986278 GQ986279 GQ986280 ADI82464.1 GQ986243 GQ986244 ADI82430.1 GQ986241 GQ986242 ADI82428.1 JN898201 AET96854.1 JN898248 AET96901.1 JN898205 AET96858.1 JN898242 AET96895.1 JN898200 AET96853.1 JN898197 JN898206 AET96859.1 GQ986245 GQ986246 ADI82432.1 JN898246 AET96899.1 JN898240 AET96893.1 GQ986249 GQ986250 GQ986260 ADI82436.1 JN898252 AET96905.1 JN897829 JN897831 AET96484.1 JN898222 AET96875.1 JN897791 AET96444.1 JN897786 AET96439.1 JN897815 AET96468.1 JN897835 AET96488.1 JN897836 JN897839 AET96492.1 JN897806 AET96459.1 JN897845 AET96498.1 JN897774 AET96427.1 JN897848 AET96501.1 JN897825 JN897826 AET96478.1 JN897804 AET96457.1 JN897801 AET96454.1
PCG77172.1 ODYU01012011 SOQ58079.1 KQ459053 KPJ04047.1 AP017484 BBB06740.1 KQ461073 KPJ09562.1 GAIX01005594 JAA86966.1 AGBW02004849 OWR55364.1 GQ986284 ADI82471.1 GQ986253 GQ986263 ADI82440.1 GQ986267 GQ986269 GQ986270 GQ986271 GQ986273 GQ986274 GQ986275 GQ986281 GQ986283 GQ986285 GQ986286 ADI82454.1 GQ986227 GQ986230 GQ986231 GQ986232 GQ986233 GQ986235 GQ986237 GQ986238 GQ986239 GQ986240 ADI82414.1 GQ986251 GQ986255 ADI82438.1 GQ986229 GQ986234 ADI82416.1 GQ986236 ADI82423.1 GQ986247 GQ986248 GQ986252 GQ986254 GQ986257 GQ986258 GQ986261 GQ986262 GQ986264 ADI82434.1 JN898214 AET96867.1 JN898213 AET96866.1 GQ986228 ADI82415.1 JN898228 AET96881.1 JN898236 AET96889.1 JN898223 AET96876.1 JN898198 JN898224 AET96851.1 JN898199 JN898208 AET96852.1 JN897799 AET96452.1 GQ986268 GQ986272 GQ986276 GQ986282 ADI82455.1 JN897820 AET96473.1 JN897847 AET96500.1 JN898233 AET96886.1 JN898202 AET96855.1 JN898207 JN898250 AET96860.1 JN898235 JN898238 AET96891.1 JN898239 JN898241 JN898243 AET96894.1 JN898251 AET96904.1 JN898211 JN898249 AET96864.1 JN898192 JN898193 AET96846.1 JN898244 JN898247 AET96897.1 JN897809 AET96462.1 JN897838 AET96491.1 JN897784 AET96437.1 JN897828 AET96481.1 JN898215 AET96868.1 JN897832 AET96485.1 JN897830 AET96483.1 JN897811 AET96464.1 JN897805 AET96458.1 JN897817 AET96470.1 JN897849 AET96502.1 JN897823 JN897824 AET96477.1 JN897798 JN897802 AET96451.1 JN897783 JN897795 JN897796 JN897797 AET96450.1 JN897785 JN897787 JN897788 JN897792 JN897793 JN897794 JN897810 JN897840 JN897842 AET96446.1 JN897821 JN897822 AET96474.1 JN897775 JN897846 AET96499.1 JN898232 AET96885.1 JN897827 AET96480.1 JN897800 AET96453.1 JN897789 AET96442.1 JN897818 AET96471.1 JN897819 AET96472.1 JN898216 AET96869.1 GQ986256 GQ986265 GQ986266 ADI82443.1 JN897816 AET96469.1 JN897814 AET96467.1 JN897773 AET96426.1 JN898210 AET96863.1 JN898217 JN898218 JN898219 JN898220 JN898221 AET96872.1 JN898225 JN898226 JN898227 AET96878.1 JN898191 JN898194 AET96847.1 JN898245 AET96898.1 GQ986259 ADI82446.1 GQ986277 GQ986278 GQ986279 GQ986280 ADI82464.1 GQ986243 GQ986244 ADI82430.1 GQ986241 GQ986242 ADI82428.1 JN898201 AET96854.1 JN898248 AET96901.1 JN898205 AET96858.1 JN898242 AET96895.1 JN898200 AET96853.1 JN898197 JN898206 AET96859.1 GQ986245 GQ986246 ADI82432.1 JN898246 AET96899.1 JN898240 AET96893.1 GQ986249 GQ986250 GQ986260 ADI82436.1 JN898252 AET96905.1 JN897829 JN897831 AET96484.1 JN898222 AET96875.1 JN897791 AET96444.1 JN897786 AET96439.1 JN897815 AET96468.1 JN897835 AET96488.1 JN897836 JN897839 AET96492.1 JN897806 AET96459.1 JN897845 AET96498.1 JN897774 AET96427.1 JN897848 AET96501.1 JN897825 JN897826 AET96478.1 JN897804 AET96457.1 JN897801 AET96454.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF69572
SSF69572
ProteinModelPortal
Q1HPK7
A0A0L7LCH9
A0A2A4K0K2
A0A2H1WYD2
A0A194QEZ3
A0A2Z5U813
+ More
A0A194QVR4 S4P5E5 A0A212FNP5 D8VH47 D8VH16 D8VH30 D8VGZ0 D8VH14 D8VGZ2 D8VGZ9 D8VH10 G9JR83 G9JR82 D8VGZ1 G9JR97 G9JRA5 G9JR92 G9JR67 G9JR68 G9JQ18 D8VH31 G9JQ39 G9JQ66 G9JRA2 G9JR71 G9JR76 G9JRA7 G9JRB0 G9JRC0 G9JR80 G9JR62 G9JRB3 G9JQ28 G9JQ57 G9JQ03 G9JQ47 G9JR84 G9JQ51 G9JQ49 G9JQ30 G9JQ24 G9JQ36 G9JQ68 G9JQ43 G9JQ17 G9JQ16 G9JQ12 G9JQ40 G9JQ65 G9JRA1 G9JQ46 G9JQ19 G9JQ08 G9JQ37 G9JQ38 G9JR85 D8VH19 G9JQ35 G9JQ33 G9JPZ2 G9JR79 G9JR88 G9JR94 G9JR63 G9JRB4 D8VH22 D8VH40 D8VH06 D8VH04 G9JR70 G9JRB7 G9JR74 G9JRB1 G9JR69 G9JR75 D8VH08 G9JRB5 G9JRA9 D8VH12 G9JRC1 G9JQ50 G9JR91 G9JQ10 G9JQ05 G9JQ34 G9JQ54 G9JQ58 G9JQ25 G9JQ64 G9JPZ3 G9JQ67 G9JQ44 G9JQ23 G9JQ20
A0A194QVR4 S4P5E5 A0A212FNP5 D8VH47 D8VH16 D8VH30 D8VGZ0 D8VH14 D8VGZ2 D8VGZ9 D8VH10 G9JR83 G9JR82 D8VGZ1 G9JR97 G9JRA5 G9JR92 G9JR67 G9JR68 G9JQ18 D8VH31 G9JQ39 G9JQ66 G9JRA2 G9JR71 G9JR76 G9JRA7 G9JRB0 G9JRC0 G9JR80 G9JR62 G9JRB3 G9JQ28 G9JQ57 G9JQ03 G9JQ47 G9JR84 G9JQ51 G9JQ49 G9JQ30 G9JQ24 G9JQ36 G9JQ68 G9JQ43 G9JQ17 G9JQ16 G9JQ12 G9JQ40 G9JQ65 G9JRA1 G9JQ46 G9JQ19 G9JQ08 G9JQ37 G9JQ38 G9JR85 D8VH19 G9JQ35 G9JQ33 G9JPZ2 G9JR79 G9JR88 G9JR94 G9JR63 G9JRB4 D8VH22 D8VH40 D8VH06 D8VH04 G9JR70 G9JRB7 G9JR74 G9JRB1 G9JR69 G9JR75 D8VH08 G9JRB5 G9JRA9 D8VH12 G9JRC1 G9JQ50 G9JR91 G9JQ10 G9JQ05 G9JQ34 G9JQ54 G9JQ58 G9JQ25 G9JQ64 G9JPZ3 G9JQ67 G9JQ44 G9JQ23 G9JQ20
PDB
6CWZ
E-value=1.0583e-74,
Score=711
Ontologies
GO
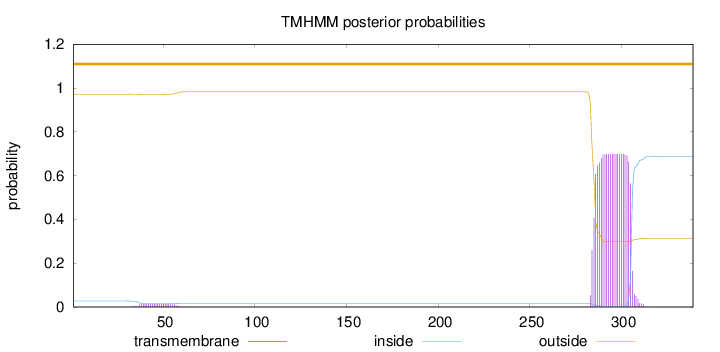
Topology
Length:
339
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
14.98651
Exp number, first 60 AAs:
0.29665
Total prob of N-in:
0.02786
outside
1 - 339
Population Genetic Test Statistics
Pi
66.941314
Theta
80.396937
Tajima's D
-1.367198
CLR
0.307916
CSRT
0.0813959302034898
Interpretation
Uncertain
