Gene
KWMTBOMO11275
Pre Gene Modal
BGIBMGA002762
Annotation
PREDICTED:_DNA_replication_licensing_factor_MCM4_[Amyelois_transitella]
Full name
DNA helicase
+ More
DNA replication licensing factor MCM4
DNA replication licensing factor mcm4-A
DNA replication licensing factor mcm4
DNA replication licensing factor mcm4-B
DNA replication licensing factor MCM4
DNA replication licensing factor mcm4-A
DNA replication licensing factor mcm4
DNA replication licensing factor mcm4-B
Alternative Name
Protein disc proliferation abnormal
CDC21 homolog-A
Minichromosome maintenance protein 4-A
P1-CDC21-A
p98
Minichromosome maintenance protein 4
CDC21 homolog-B
Minichromosome maintenance protein 4-B
P1-CDC21-B
CDC21 homolog-A
Minichromosome maintenance protein 4-A
P1-CDC21-A
p98
Minichromosome maintenance protein 4
CDC21 homolog-B
Minichromosome maintenance protein 4-B
P1-CDC21-B
Location in the cell
Nuclear Reliability : 2.169
Sequence
CDS
ATGTCTACACCATCTAAACGCCCGTCGTCGCGTGCATCATCGGAAGCATCGACTCCACGAAGAACGCGATCCTCACAACAAGTTATCGTTCCGGAGACTCCGCAGCGTTCAACCGGCACACCTTCGAAAGATGGCACTCCAAGTACACCACGAGCAGCTCCCGGTCCATCTCGGACACCCCGTACACCTCGTGAAGCTCCAGGAACATCACCTGCTCGAGCTCTTGCATCCAGTCCCATAGGCACAGATATAGACATGAGCTCTCCTCTTAATTACGGCACTCCAAGTTCTCTAAGCACTCCTCGCTCGCTCATGAGAGGCATCATGACCCCAGCACGACAGAGGGCTGACCTCCGAGGTCACCATATAGCTCCCAATGTACCGGCACCCACACCTACACGGAGAGTGGAGGGTTCAGAGGAGGCTTCGAGTGAGCCGCAGCTGGTCGTGTGGGGGACGGACGTGGCCATCGCAGAATGCAGGGAGAAATTCATCAAGTTCATCCAGAGATACGTGGAGCCAACTGCTGTCACCAACGAACCTCTTTACGAGCAAAAACTTGAGGAGATTCACACATTGGAAGAGCCATTCTTAGATGTGGACTGTGAACATGTCAGAGTTTTCGATTCCAAACTACACAGACAACTGGTCTCTTATCCCCAAGAGGTTGTGCCGGCATTCGACGCAGCAGTCAACGAGCTGTTCTTCGAGAAGTACCCGGCGGCCGTGCTCGAGCACCAGATCCAGGTCCGGCCGTACAACGCCCCGCCGCGACACATGAGAGACCTCAATCCGGAAGACATAGACCAGCTGGTGACTATATCGGGCATGGTGATACGCACGTCCGGCATCGTGCCGGAGATGCGGGAGGCGTTCTTCAAGTGCGCGGTGTGCGGCGCCGCGGCGGGCGGGGAGCTGCAGCGCGGCCGCGTGCCCGAGCCCGCACACTGCGCGCACTGCAACACGGCGCACTGCTTCCAGCTCATACACAACCGCTCGCACTTCAGCGACAAGCAGCTCGTCAAGCTGCAGGAGGCACCTGGTGACTGGTTTTTTTATTTTATTTTTATTGCTTAG
Protein
MSTPSKRPSSRASSEASTPRRTRSSQQVIVPETPQRSTGTPSKDGTPSTPRAAPGPSRTPRTPREAPGTSPARALASSPIGTDIDMSSPLNYGTPSSLSTPRSLMRGIMTPARQRADLRGHHIAPNVPAPTPTRRVEGSEEASSEPQLVVWGTDVAIAECREKFIKFIQRYVEPTAVTNEPLYEQKLEEIHTLEEPFLDVDCEHVRVFDSKLHRQLVSYPQEVVPAFDAAVNELFFEKYPAAVLEHQIQVRPYNAPPRHMRDLNPEDIDQLVTISGMVIRTSGIVPEMREAFFKCAVCGAAAGGELQRGRVPEPAHCAHCNTAHCFQLIHNRSHFSDKQLVKLQEAPGDWFFYFIFIA
Summary
Description
Acts as component of the Mcm2-7 complex (Mcm complex) which is the putative replicative helicase essential for 'once per cell cycle' DNA replication initiation and elongation in eukaryotic cells. The active ATPase sites in the Mcm2-7 ring are formed through the interaction surfaces of two neighboring subunits such that a critical structure of a conserved arginine finger motif is provided in trans relative to the ATP-binding site of the Walker A box of the adjacent subunit. The six ATPase active sites, however, are likely to contribute differentially to the complex helicase activity. Required for DNA replication and cell proliferation. Essential role in mitotic DNA replication but not in endoreplication.
Acts as component of the mcm2-7 complex (mcm complex) which is the putative replicative helicase essential for 'once per cell cycle' DNA replication initiation and elongation in eukaryotic cells. The active ATPase sites in the mcm2-7 ring are formed through the interaction surfaces of two neighboring subunits such that a critical structure of a conserved arginine finger motif is provided in trans relative to the ATP-binding site of the Walker A box of the adjacent subunit. The six ATPase active sites, however, are likely to contribute differentially to the complex helicase activity.
Acts as component of the mcm2-7 complex (mcm complex) which is the putative replicative helicase essential for 'once per cell cycle' DNA replication initiation and elongation in eukaryotic cells. The active ATPase sites in the mcm2-7 ring are formed through the interaction surfaces of two neighboring subunits such that a critical structure of a conserved arginine finger motif is provided in trans relative to the ATP-binding site of the Walker A box of the adjacent subunit. The six ATPase active sites, however, are likely to contribute differentially to the complex helicase activity.
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Subunit
Component of the Mcm2-7 complex. The complex forms a toroidal hexameric ring with the proposed subunit order Mcm2-Mcm6-Mcm4-Mcm7-Mcm3-Mcm5 (Probable).
Component of the mcm2-7 complex (RLF-M). The complex forms a toroidal hexameric ring with the proposed subunit order mcm2-mcm6-mcm4-mcm7-mcm3-mcm5 (Probable). The heterodimer of mmcm3/mcm5 interacts with mcm4, mmcm6, mcm7 and weakly with mcm2.
Component of the mcm2-7 complex (RLF-M). The complex forms a toroidal hexameric ring with the proposed subunit order mcm2-mcm6-mcm4-mcm7-mcm3-mcm5 (Probable). The heterodimer of mmcm3/mcm5 interacts with mcm4, mmcm6, mcm7 and weakly with mcm2. Begins to associate with zmcm6 at the neurula stage (By similarity).
Component of the mcm2-7 complex (RLF-M). The complex forms a toroidal hexameric ring with the proposed subunit order mcm2-mcm6-mcm4-mcm7-mcm3-mcm5 (Probable). The heterodimer of mmcm3/mcm5 interacts with mcm4, mmcm6, mcm7 and weakly with mcm2. Begins to associate with zmcm6 at the neurula stage.
Component of the mcm2-7 complex (RLF-M). The complex forms a toroidal hexameric ring with the proposed subunit order mcm2-mcm6-mcm4-mcm7-mcm3-mcm5 (Probable). The heterodimer of mmcm3/mcm5 interacts with mcm4, mmcm6, mcm7 and weakly with mcm2.
Component of the mcm2-7 complex (RLF-M). The complex forms a toroidal hexameric ring with the proposed subunit order mcm2-mcm6-mcm4-mcm7-mcm3-mcm5 (Probable). The heterodimer of mmcm3/mcm5 interacts with mcm4, mmcm6, mcm7 and weakly with mcm2. Begins to associate with zmcm6 at the neurula stage (By similarity).
Component of the mcm2-7 complex (RLF-M). The complex forms a toroidal hexameric ring with the proposed subunit order mcm2-mcm6-mcm4-mcm7-mcm3-mcm5 (Probable). The heterodimer of mmcm3/mcm5 interacts with mcm4, mmcm6, mcm7 and weakly with mcm2. Begins to associate with zmcm6 at the neurula stage.
Similarity
Belongs to the MCM family.
Keywords
ATP-binding
Complete proteome
DNA replication
DNA-binding
Helicase
Hydrolase
Nucleotide-binding
Nucleus
Phosphoprotein
Reference proteome
Cell cycle
Metal-binding
Zinc
Zinc-finger
Feature
chain DNA replication licensing factor MCM4
Uniprot
A0A212FER1
A0A2H1WYC1
A0A2A4K0L3
A0A194QX82
A0A194QF49
A0A182H304
+ More
Q16TZ0 Q5BI95 A0A1Q3EVE7 A0A0K8WM27 A0A034W864 B0WVG5 A0A1I8QAN5 A0A1I8N9V2 A0A1A9USP5 A0A0L0C837 B3N9Y3 B4P1Q4 B3MGV3 A0A0A1XAF5 A0A182VI32 A0A182TDY8 W8C9Q3 A0A0J9TVC7 B4HQV7 Q7PPI5 A0A182XEH1 A0A182KNP9 A0A182HQL4 A0A182ILA0 A0A0B4LEV2 Q26454 B4KQ97 A0A1W4VDK0 A0A182N1Y6 A0A182T370 B4LLC4 A0A0C9QKK7 A0A182WM22 A0A1W4XPI5 A0A182RE53 V5KX69 A0A182PTV7 K7INC3 A0A0M3QUM2 A0A182M1N7 Q28YI1 A0A2P8XCP1 A0A067R0Z3 B4MIN6 A0A084WE11 A0A1B0G5U6 B4K2W4 W5JWV7 A0A3B0J6N2 A0A182JVZ5 A0A1B6KRG0 A0A1B6F0W3 A0A1Y1KIS5 T1P9F1 B4H8K8 A0A182YR51 B4J9Z6 A0A182FLN8 A0A232EYL2 D6WSI2 A0A182Q843 N6THZ9 A0A0J7L4H7 A0A2J7PJ85 A0A026WMZ8 A0A336K914 A0A3R7PR63 A0A1J1HY95 A0A2I4CZX7 A0A3Q2ZSX3 A0A1W5A084 A0A3B3RBL6 A0A1S3LXE0 Q5XK83 A0A1L8FSY2 A0A3Q1HY34 A0A060W8M7 Q6GL41 E2A9C4 E2C0X5 V4A2X0 A0A2P4SU88 A0A146YY99 C0HAN5 A0A3Q2E9N7 A0A2U9BJR0 V8P321 A0A2D4MKN7 A0A3P9AIQ8 A0A3B4V4A7 A0A1L8FYX1 P30664 A0A091R783
Q16TZ0 Q5BI95 A0A1Q3EVE7 A0A0K8WM27 A0A034W864 B0WVG5 A0A1I8QAN5 A0A1I8N9V2 A0A1A9USP5 A0A0L0C837 B3N9Y3 B4P1Q4 B3MGV3 A0A0A1XAF5 A0A182VI32 A0A182TDY8 W8C9Q3 A0A0J9TVC7 B4HQV7 Q7PPI5 A0A182XEH1 A0A182KNP9 A0A182HQL4 A0A182ILA0 A0A0B4LEV2 Q26454 B4KQ97 A0A1W4VDK0 A0A182N1Y6 A0A182T370 B4LLC4 A0A0C9QKK7 A0A182WM22 A0A1W4XPI5 A0A182RE53 V5KX69 A0A182PTV7 K7INC3 A0A0M3QUM2 A0A182M1N7 Q28YI1 A0A2P8XCP1 A0A067R0Z3 B4MIN6 A0A084WE11 A0A1B0G5U6 B4K2W4 W5JWV7 A0A3B0J6N2 A0A182JVZ5 A0A1B6KRG0 A0A1B6F0W3 A0A1Y1KIS5 T1P9F1 B4H8K8 A0A182YR51 B4J9Z6 A0A182FLN8 A0A232EYL2 D6WSI2 A0A182Q843 N6THZ9 A0A0J7L4H7 A0A2J7PJ85 A0A026WMZ8 A0A336K914 A0A3R7PR63 A0A1J1HY95 A0A2I4CZX7 A0A3Q2ZSX3 A0A1W5A084 A0A3B3RBL6 A0A1S3LXE0 Q5XK83 A0A1L8FSY2 A0A3Q1HY34 A0A060W8M7 Q6GL41 E2A9C4 E2C0X5 V4A2X0 A0A2P4SU88 A0A146YY99 C0HAN5 A0A3Q2E9N7 A0A2U9BJR0 V8P321 A0A2D4MKN7 A0A3P9AIQ8 A0A3B4V4A7 A0A1L8FYX1 P30664 A0A091R783
EC Number
3.6.4.12
Pubmed
22118469
26354079
26483478
17510324
25348373
25315136
+ More
26108605 17994087 17550304 25830018 24495485 22936249 12364791 14747013 17210077 20966253 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 7489728 16798881 17372656 18327897 20122406 20075255 15632085 29403074 24845553 24438588 20920257 23761445 28004739 25244985 28648823 18362917 19820115 23537049 24508170 30249741 29240929 9214647 9214646 16204181 27762356 24755649 20798317 23254933 20433749 24297900 25069045 8605878 8901561 1454522 9512418 9851868 10779356 16369567
26108605 17994087 17550304 25830018 24495485 22936249 12364791 14747013 17210077 20966253 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 7489728 16798881 17372656 18327897 20122406 20075255 15632085 29403074 24845553 24438588 20920257 23761445 28004739 25244985 28648823 18362917 19820115 23537049 24508170 30249741 29240929 9214647 9214646 16204181 27762356 24755649 20798317 23254933 20433749 24297900 25069045 8605878 8901561 1454522 9512418 9851868 10779356 16369567
EMBL
AGBW02008904
OWR52231.1
ODYU01012011
SOQ58075.1
NWSH01000348
PCG77182.1
+ More
KQ461073 KPJ09565.1 KQ459053 KPJ04044.1 JXUM01024334 JXUM01024335 JXUM01087698 JXUM01087699 KQ563653 KQ560687 KXJ73554.1 KXJ81271.1 CH477637 EAT37976.1 BT021329 AAX33477.1 GFDL01015765 JAV19280.1 GDHF01000205 JAI52109.1 GAKP01007211 JAC51741.1 DS232126 EDS35571.1 JRES01000780 KNC28392.1 CH954177 EDV59679.1 CM000157 EDW89190.1 CH902619 EDV37871.1 GBXI01006639 JAD07653.1 GAMC01001959 JAC04597.1 CM002911 KMY92000.1 CH480816 EDW46767.1 AAAB01008948 EAA10355.3 APCN01004657 AE013599 AHN55958.1 S80255 CH933808 EDW09225.1 CH940648 EDW61876.1 GBYB01015313 JAG85080.1 KF471133 AHA42533.1 CP012524 ALC40873.1 AXCM01003001 CM000071 EAL25984.1 PYGN01002995 PSN29765.1 KK852792 KDR16461.1 CH963719 EDW71975.1 ATLV01023101 KE525340 KFB48455.1 CCAG010007472 CH920114 EDV90433.1 ADMH02000108 ETN67810.1 OUUW01000001 SPP75703.1 GEBQ01025966 JAT14011.1 GECZ01025975 JAS43794.1 GEZM01085192 JAV60140.1 KA645406 AFP60035.1 CH479223 EDW35043.1 CH916367 EDW02583.1 NNAY01001603 OXU23451.1 KQ971352 EFA06648.2 AXCN02000977 APGK01025879 KB740605 ENN80029.1 LBMM01000833 KMQ97399.1 NEVH01024950 PNF16400.1 KK107151 QOIP01000003 EZA57318.1 RLU25202.1 UFQS01000188 UFQT01000188 SSX00981.1 SSX21361.1 QCYY01001161 ROT80053.1 CVRI01000030 CRK92530.1 U44049 BC083031 AAC60225.1 AAH83031.1 CM004477 OCT74714.1 FR904442 CDQ63477.1 BC074670 AAH74670.1 GL437781 EFN69966.1 GL451850 EFN78411.1 KB201262 ESO98208.1 PPHD01022692 POI27685.1 GCES01027262 JAR59061.1 BT059391 ACN11104.1 CP026249 AWP03956.1 AZIM01000957 ETE68700.1 IACM01111057 LAB33628.1 CM004476 OCT76788.1 U29178 U46131 BC072870 Z15033 KK714268 KFQ35316.1
KQ461073 KPJ09565.1 KQ459053 KPJ04044.1 JXUM01024334 JXUM01024335 JXUM01087698 JXUM01087699 KQ563653 KQ560687 KXJ73554.1 KXJ81271.1 CH477637 EAT37976.1 BT021329 AAX33477.1 GFDL01015765 JAV19280.1 GDHF01000205 JAI52109.1 GAKP01007211 JAC51741.1 DS232126 EDS35571.1 JRES01000780 KNC28392.1 CH954177 EDV59679.1 CM000157 EDW89190.1 CH902619 EDV37871.1 GBXI01006639 JAD07653.1 GAMC01001959 JAC04597.1 CM002911 KMY92000.1 CH480816 EDW46767.1 AAAB01008948 EAA10355.3 APCN01004657 AE013599 AHN55958.1 S80255 CH933808 EDW09225.1 CH940648 EDW61876.1 GBYB01015313 JAG85080.1 KF471133 AHA42533.1 CP012524 ALC40873.1 AXCM01003001 CM000071 EAL25984.1 PYGN01002995 PSN29765.1 KK852792 KDR16461.1 CH963719 EDW71975.1 ATLV01023101 KE525340 KFB48455.1 CCAG010007472 CH920114 EDV90433.1 ADMH02000108 ETN67810.1 OUUW01000001 SPP75703.1 GEBQ01025966 JAT14011.1 GECZ01025975 JAS43794.1 GEZM01085192 JAV60140.1 KA645406 AFP60035.1 CH479223 EDW35043.1 CH916367 EDW02583.1 NNAY01001603 OXU23451.1 KQ971352 EFA06648.2 AXCN02000977 APGK01025879 KB740605 ENN80029.1 LBMM01000833 KMQ97399.1 NEVH01024950 PNF16400.1 KK107151 QOIP01000003 EZA57318.1 RLU25202.1 UFQS01000188 UFQT01000188 SSX00981.1 SSX21361.1 QCYY01001161 ROT80053.1 CVRI01000030 CRK92530.1 U44049 BC083031 AAC60225.1 AAH83031.1 CM004477 OCT74714.1 FR904442 CDQ63477.1 BC074670 AAH74670.1 GL437781 EFN69966.1 GL451850 EFN78411.1 KB201262 ESO98208.1 PPHD01022692 POI27685.1 GCES01027262 JAR59061.1 BT059391 ACN11104.1 CP026249 AWP03956.1 AZIM01000957 ETE68700.1 IACM01111057 LAB33628.1 CM004476 OCT76788.1 U29178 U46131 BC072870 Z15033 KK714268 KFQ35316.1
Proteomes
UP000007151
UP000218220
UP000053240
UP000053268
UP000069940
UP000249989
+ More
UP000008820 UP000002320 UP000095300 UP000095301 UP000078200 UP000037069 UP000008711 UP000002282 UP000007801 UP000075903 UP000075902 UP000001292 UP000007062 UP000076407 UP000075882 UP000075840 UP000075880 UP000000803 UP000009192 UP000192221 UP000075884 UP000075901 UP000008792 UP000075920 UP000192223 UP000075900 UP000075885 UP000002358 UP000092553 UP000075883 UP000001819 UP000245037 UP000027135 UP000007798 UP000030765 UP000092444 UP000001070 UP000000673 UP000268350 UP000075881 UP000008744 UP000076408 UP000069272 UP000215335 UP000007266 UP000075886 UP000019118 UP000036403 UP000235965 UP000053097 UP000279307 UP000283509 UP000183832 UP000192220 UP000264800 UP000192224 UP000261540 UP000087266 UP000186698 UP000265040 UP000193380 UP000008143 UP000000311 UP000008237 UP000030746 UP000265020 UP000246464 UP000265140 UP000261420
UP000008820 UP000002320 UP000095300 UP000095301 UP000078200 UP000037069 UP000008711 UP000002282 UP000007801 UP000075903 UP000075902 UP000001292 UP000007062 UP000076407 UP000075882 UP000075840 UP000075880 UP000000803 UP000009192 UP000192221 UP000075884 UP000075901 UP000008792 UP000075920 UP000192223 UP000075900 UP000075885 UP000002358 UP000092553 UP000075883 UP000001819 UP000245037 UP000027135 UP000007798 UP000030765 UP000092444 UP000001070 UP000000673 UP000268350 UP000075881 UP000008744 UP000076408 UP000069272 UP000215335 UP000007266 UP000075886 UP000019118 UP000036403 UP000235965 UP000053097 UP000279307 UP000283509 UP000183832 UP000192220 UP000264800 UP000192224 UP000261540 UP000087266 UP000186698 UP000265040 UP000193380 UP000008143 UP000000311 UP000008237 UP000030746 UP000265020 UP000246464 UP000265140 UP000261420
Interpro
IPR008047
MCM_4
+ More
IPR018525 MCM_CS
IPR027925 MCM_N
IPR041562 MCM_lid
IPR027417 P-loop_NTPase
IPR033762 MCM_OB
IPR031327 MCM
IPR001208 MCM_dom
IPR012340 NA-bd_OB-fold
IPR003593 AAA+_ATPase
IPR001005 SANT/Myb
IPR017884 SANT_dom
IPR009057 Homeobox-like_sf
IPR000949 ELM2_dom
IPR004210 BESS_motif
IPR006578 MADF-dom
IPR018525 MCM_CS
IPR027925 MCM_N
IPR041562 MCM_lid
IPR027417 P-loop_NTPase
IPR033762 MCM_OB
IPR031327 MCM
IPR001208 MCM_dom
IPR012340 NA-bd_OB-fold
IPR003593 AAA+_ATPase
IPR001005 SANT/Myb
IPR017884 SANT_dom
IPR009057 Homeobox-like_sf
IPR000949 ELM2_dom
IPR004210 BESS_motif
IPR006578 MADF-dom
ProteinModelPortal
A0A212FER1
A0A2H1WYC1
A0A2A4K0L3
A0A194QX82
A0A194QF49
A0A182H304
+ More
Q16TZ0 Q5BI95 A0A1Q3EVE7 A0A0K8WM27 A0A034W864 B0WVG5 A0A1I8QAN5 A0A1I8N9V2 A0A1A9USP5 A0A0L0C837 B3N9Y3 B4P1Q4 B3MGV3 A0A0A1XAF5 A0A182VI32 A0A182TDY8 W8C9Q3 A0A0J9TVC7 B4HQV7 Q7PPI5 A0A182XEH1 A0A182KNP9 A0A182HQL4 A0A182ILA0 A0A0B4LEV2 Q26454 B4KQ97 A0A1W4VDK0 A0A182N1Y6 A0A182T370 B4LLC4 A0A0C9QKK7 A0A182WM22 A0A1W4XPI5 A0A182RE53 V5KX69 A0A182PTV7 K7INC3 A0A0M3QUM2 A0A182M1N7 Q28YI1 A0A2P8XCP1 A0A067R0Z3 B4MIN6 A0A084WE11 A0A1B0G5U6 B4K2W4 W5JWV7 A0A3B0J6N2 A0A182JVZ5 A0A1B6KRG0 A0A1B6F0W3 A0A1Y1KIS5 T1P9F1 B4H8K8 A0A182YR51 B4J9Z6 A0A182FLN8 A0A232EYL2 D6WSI2 A0A182Q843 N6THZ9 A0A0J7L4H7 A0A2J7PJ85 A0A026WMZ8 A0A336K914 A0A3R7PR63 A0A1J1HY95 A0A2I4CZX7 A0A3Q2ZSX3 A0A1W5A084 A0A3B3RBL6 A0A1S3LXE0 Q5XK83 A0A1L8FSY2 A0A3Q1HY34 A0A060W8M7 Q6GL41 E2A9C4 E2C0X5 V4A2X0 A0A2P4SU88 A0A146YY99 C0HAN5 A0A3Q2E9N7 A0A2U9BJR0 V8P321 A0A2D4MKN7 A0A3P9AIQ8 A0A3B4V4A7 A0A1L8FYX1 P30664 A0A091R783
Q16TZ0 Q5BI95 A0A1Q3EVE7 A0A0K8WM27 A0A034W864 B0WVG5 A0A1I8QAN5 A0A1I8N9V2 A0A1A9USP5 A0A0L0C837 B3N9Y3 B4P1Q4 B3MGV3 A0A0A1XAF5 A0A182VI32 A0A182TDY8 W8C9Q3 A0A0J9TVC7 B4HQV7 Q7PPI5 A0A182XEH1 A0A182KNP9 A0A182HQL4 A0A182ILA0 A0A0B4LEV2 Q26454 B4KQ97 A0A1W4VDK0 A0A182N1Y6 A0A182T370 B4LLC4 A0A0C9QKK7 A0A182WM22 A0A1W4XPI5 A0A182RE53 V5KX69 A0A182PTV7 K7INC3 A0A0M3QUM2 A0A182M1N7 Q28YI1 A0A2P8XCP1 A0A067R0Z3 B4MIN6 A0A084WE11 A0A1B0G5U6 B4K2W4 W5JWV7 A0A3B0J6N2 A0A182JVZ5 A0A1B6KRG0 A0A1B6F0W3 A0A1Y1KIS5 T1P9F1 B4H8K8 A0A182YR51 B4J9Z6 A0A182FLN8 A0A232EYL2 D6WSI2 A0A182Q843 N6THZ9 A0A0J7L4H7 A0A2J7PJ85 A0A026WMZ8 A0A336K914 A0A3R7PR63 A0A1J1HY95 A0A2I4CZX7 A0A3Q2ZSX3 A0A1W5A084 A0A3B3RBL6 A0A1S3LXE0 Q5XK83 A0A1L8FSY2 A0A3Q1HY34 A0A060W8M7 Q6GL41 E2A9C4 E2C0X5 V4A2X0 A0A2P4SU88 A0A146YY99 C0HAN5 A0A3Q2E9N7 A0A2U9BJR0 V8P321 A0A2D4MKN7 A0A3P9AIQ8 A0A3B4V4A7 A0A1L8FYX1 P30664 A0A091R783
PDB
6HV9
E-value=4.83071e-32,
Score=344
Ontologies
GO
GO:0042555
GO:0005524
GO:0003678
GO:0003677
GO:0005634
GO:0006270
GO:0043138
GO:0007052
GO:0003697
GO:0006268
GO:0006271
GO:0006267
GO:0000727
GO:0003688
GO:1902975
GO:0006260
GO:0004386
GO:0003682
GO:0001878
GO:0007049
GO:0046872
GO:0030174
GO:0000785
GO:0015935
GO:0006811
GO:0005622
GO:0006810
GO:0003824
PANTHER
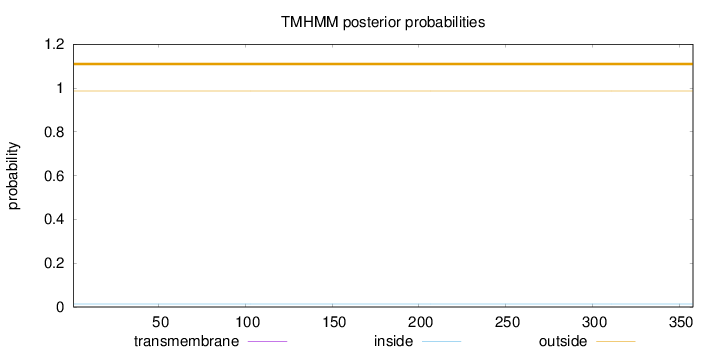
Topology
Subcellular location
Nucleus
Length:
358
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00126
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01388
outside
1 - 358
Population Genetic Test Statistics
Pi
288.645183
Theta
171.762322
Tajima's D
1.794012
CLR
0.411089
CSRT
0.846207689615519
Interpretation
Uncertain
