Gene
KWMTBOMO11271 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002759
Annotation
PREDICTED:_ubiquitin_carboxyl-terminal_hydrolase_14_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 1.173 Nuclear Reliability : 1.674
Sequence
CDS
ATGCCTAACGTTTCAGTGAAGGTAAAATGGGGCAAGGAAATGTACCCAGACGTGGAGGTGAATACAGACGATGAACCGGTACTGTTCAAGGCGCAGATATTTGCACTGACAGGGGTCCAGCCAGAGAGACAAAAAGTCGTTTGCAAAGGAGTCACATTACGGGATGACACATGGGGCAATTTTAAATTGACGAATAACGCCTTAGTACTTGTCATGGGCAGTAAAGAGGAAGACGTGCCAGCAGCACCAGTGGAACAGACACGTTTTGTCGAAGACATGAATGAAGCTGAACTGGCTACAGCCATGGATATGCCAGAGGGTCTCATCAACTTGGGTAACACTTGTTACATGAATGCGACAGTGCAGTGTCTGAAGACGGTCCCTGAACTTAGAAATGCCTTGCTCAATTATGATAAATCATCAACGGGCGGCGACACAGCGGGAGAACTAACGGCGGCACTAAGCGAGACGGTCTCGGCCCTGGACTCCGGGGGCTCCAGCGCGTGCGGCGTGGCGGCGGCGCGCCTGCTGGGAGCCCTGCACGGCGCGGCGCCTCGCTTCGCGGAGCGGGCGGCCGCGGGGGGCTTCGCGCAGCAGGACGCCTCCGAGTGCTGGACCGAAATCGTGAGGGCGTTGCAGCAGAGGCTCCCGGCCGCACCCATGCCAGATAAGAACGCGACAAGGGCATCAGTGATCGAGCAATACTTCGGAGGTACCCTGGATGTGGAGCTTGTATGCTCAGAGGCTGATGAGCCCCCGACCCACTCTGAGGAGTCATTCCTCCAGTTGTCCTGTTTCATATCACAAGACGTCAAATATCTGCAGTCTGGACTTAGATCTAAAATGGCTGAGCATATAACAAAGATGTCACAAACACTGGGCAGGGATGCTGTGTACACAAAAACTAGTAAAATAAGCCGTCTGCCTGCTTATTTGACAGTGCAATTCGTTCGTTTCTACTACAAGGAGAAAGAATCTATCAACGCTAAAATACTAAAAGATGTTAAATTCCCTCTGGACTTGGACGTGTATGAACTTTGCACACCGGAACTACAAGAGAGGCTGACACCGATGCGAAATAAATTCAAGGAGTTGGAAGATGCGAAAGTACAATCATCGCTTAGCTCCGGTAACAAGAGTCACGGGGACAGCAGCAAGGCCAAGAAGAAGCGTGTCTTGCCCTACTGGTTCGAAAACGACGTAGGCAGCAACAACAGCGGCTACTACCGGCTGCAGGCCGTGCTCACGCACCGCGGCCGCTCGTCCTCCTCCGGGCACTACGTGGCGTGGGTGGCGCGCGGCGCGGGCTGGCTGCGCTGCGACGACGACACCGTCACCCCCGTCACCGAGGACGAGGTGCTCAAGCTCAGCGGGGGAGGTGACTGGCACTGCGCCTACCTTCTGCTGTACGGACCTAAAATCCTGGAGCTCCCCGAGCAGGATGAGGACGAGCCCATGGTCACGGACATAACAGACGATCCTGCTGGGGAACCCCCCACAGCGCTCGCATAA
Protein
MPNVSVKVKWGKEMYPDVEVNTDDEPVLFKAQIFALTGVQPERQKVVCKGVTLRDDTWGNFKLTNNALVLVMGSKEEDVPAAPVEQTRFVEDMNEAELATAMDMPEGLINLGNTCYMNATVQCLKTVPELRNALLNYDKSSTGGDTAGELTAALSETVSALDSGGSSACGVAAARLLGALHGAAPRFAERAAAGGFAQQDASECWTEIVRALQQRLPAAPMPDKNATRASVIEQYFGGTLDVELVCSEADEPPTHSEESFLQLSCFISQDVKYLQSGLRSKMAEHITKMSQTLGRDAVYTKTSKISRLPAYLTVQFVRFYYKEKESINAKILKDVKFPLDLDVYELCTPELQERLTPMRNKFKELEDAKVQSSLSSGNKSHGDSSKAKKKRVLPYWFENDVGSNNSGYYRLQAVLTHRGRSSSSGHYVAWVARGAGWLRCDDDTVTPVTEDEVLKLSGGGDWHCAYLLLYGPKILELPEQDEDEPMVTDITDDPAGEPPTALA
Summary
Similarity
Belongs to the peptidase C19 family.
Uniprot
A0A194QGF7
A0A2H1WNJ8
H9IZS4
A0A194QW90
D6WPP3
A0A1B6EH15
+ More
A0A1Y1MW02 E2BDN2 A0A1B6GDA5 A0A1B6CDA9 A0A087ZQN5 A0A0L7QXV5 A0A1W4X482 A0A2A3E461 U4UIN8 A0A154PAW0 J3JZF1 A0A2J7QET0 E9J3G3 F4WPY4 A0A0N0U6N3 A0A3L8DAW3 A0A2J7QER1 A0A151JU35 A0A0C9R6V4 A0A151J194 A0A151HYU3 A0A151WGX3 A0A232FAQ5 K7IZG1 A0A0N8E077 E9H1J0 A0A026WAW6 A0A0P6AB63 A0A0P6IKD3 A0A182VAE0 A0A2L2Y6Q4 A0A182Q9F0 A0A1B6LZN6 U5EZI9 J9JTF1 A0A2M4BKR3 A0A1S4FQW4 A0A2H8TGA1 A0A087TJS9 A0A1S4H1L1 T1IX76 A0A2M4ADJ1 T1E8N2 A0A2S2P739 Q7QGE8 A0A2M4CSG8 V9IKQ5 E0VQI8 A0A0N7Z981 A0A2R5LJH8 A0A3S3RZB5 V4ALX1 A0A336K563 W8BWE7 A0A069DUM5 A0A0A1XK47 T1IFE6 A0A0P5CEX1 A0A0L0BQE2 Q29NI2 B4G7H7 A0A1Q3FVI4 A0A1S4EF74 A0A293MSB2 A0A034WU48 A0A224XNG6 A0A3B0JMS0 B4KJF9 A0A0V0G683 A0A3M6TW59 A0A0M4EC75 B4NZ95 B4Q8T6 Q9VKZ8 B4HWK5 B4LR09 A0A0K8U1E3 B3N970 V5HHU1 A0A1I8PCJ9 A0A131XU62 B3MPR0 L7M340 A0A1W4UUS3 A0A023F4P6 A0A0P4VVM3 B4JPP8 A0A1I8MRJ9 A0A1B0A536 A0A224Z2W2 A0A131YZR6 T1PCQ4 A0A1A9V9I6 D3TQ68
A0A1Y1MW02 E2BDN2 A0A1B6GDA5 A0A1B6CDA9 A0A087ZQN5 A0A0L7QXV5 A0A1W4X482 A0A2A3E461 U4UIN8 A0A154PAW0 J3JZF1 A0A2J7QET0 E9J3G3 F4WPY4 A0A0N0U6N3 A0A3L8DAW3 A0A2J7QER1 A0A151JU35 A0A0C9R6V4 A0A151J194 A0A151HYU3 A0A151WGX3 A0A232FAQ5 K7IZG1 A0A0N8E077 E9H1J0 A0A026WAW6 A0A0P6AB63 A0A0P6IKD3 A0A182VAE0 A0A2L2Y6Q4 A0A182Q9F0 A0A1B6LZN6 U5EZI9 J9JTF1 A0A2M4BKR3 A0A1S4FQW4 A0A2H8TGA1 A0A087TJS9 A0A1S4H1L1 T1IX76 A0A2M4ADJ1 T1E8N2 A0A2S2P739 Q7QGE8 A0A2M4CSG8 V9IKQ5 E0VQI8 A0A0N7Z981 A0A2R5LJH8 A0A3S3RZB5 V4ALX1 A0A336K563 W8BWE7 A0A069DUM5 A0A0A1XK47 T1IFE6 A0A0P5CEX1 A0A0L0BQE2 Q29NI2 B4G7H7 A0A1Q3FVI4 A0A1S4EF74 A0A293MSB2 A0A034WU48 A0A224XNG6 A0A3B0JMS0 B4KJF9 A0A0V0G683 A0A3M6TW59 A0A0M4EC75 B4NZ95 B4Q8T6 Q9VKZ8 B4HWK5 B4LR09 A0A0K8U1E3 B3N970 V5HHU1 A0A1I8PCJ9 A0A131XU62 B3MPR0 L7M340 A0A1W4UUS3 A0A023F4P6 A0A0P4VVM3 B4JPP8 A0A1I8MRJ9 A0A1B0A536 A0A224Z2W2 A0A131YZR6 T1PCQ4 A0A1A9V9I6 D3TQ68
Pubmed
26354079
19121390
18362917
19820115
28004739
20798317
+ More
23537049 22516182 21282665 21719571 30249741 28648823 20075255 21292972 24508170 26561354 12364791 20566863 27129103 23254933 24495485 26334808 25830018 26108605 15632085 17994087 25348373 30382153 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25765539 25576852 25474469 25315136 28797301 26830274 20353571
23537049 22516182 21282665 21719571 30249741 28648823 20075255 21292972 24508170 26561354 12364791 20566863 27129103 23254933 24495485 26334808 25830018 26108605 15632085 17994087 25348373 30382153 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25765539 25576852 25474469 25315136 28797301 26830274 20353571
EMBL
KQ459053
KPJ04040.1
ODYU01009911
SOQ54645.1
BABH01039951
BABH01039952
+ More
KQ461073 KPJ09569.1 KQ971354 EFA06193.1 GEDC01000107 JAS37191.1 GEZM01022944 JAV88645.1 GL447678 EFN86194.1 GECZ01009337 JAS60432.1 GEDC01025861 JAS11437.1 KQ414699 KOC63450.1 KZ288386 PBC26480.1 KB632319 ERL92273.1 KQ434846 KZC08320.1 APGK01045645 BT128633 KB741037 AEE63590.1 ENN74687.1 NEVH01015306 PNF27077.1 GL768066 EFZ12593.1 GL888258 EGI63735.1 KQ435724 KOX78344.1 QOIP01000010 RLU17451.1 PNF27078.1 KQ981829 KYN35072.1 GBYB01012064 JAG81831.1 KQ980541 KYN15590.1 KQ976717 KYM76785.1 KQ983140 KYQ47074.1 NNAY01000510 OXU27924.1 GDIQ01074652 LRGB01000024 JAN20085.1 KZS21373.1 GL732583 EFX74497.1 KK107348 EZA52154.1 GDIP01033149 JAM70566.1 GDIQ01009631 JAN85106.1 IAAA01003043 IAAA01003044 LAA02825.1 AXCN02000845 GEBQ01010831 JAT29146.1 GANO01000714 JAB59157.1 ABLF02040831 GGFJ01004471 MBW53612.1 GFXV01001265 MBW13070.1 KK115530 KFM65368.1 AAAB01008835 JH431644 GGFK01005471 MBW38792.1 GAMD01002613 JAA98977.1 GGMR01012650 MBY25269.1 EAA05748.4 GGFL01004114 MBW68292.1 JR052538 AEY61718.1 DS235430 EEB15644.1 GDKW01001283 JAI55312.1 GGLE01005574 MBY09700.1 NCKU01003779 NCKU01002879 NCKU01002874 RWS06929.1 RWS08623.1 RWS08633.1 KB199650 ESP05184.1 UFQS01000111 UFQT01000111 SSW99799.1 SSX20179.1 GAMC01000905 JAC05651.1 GBGD01001239 JAC87650.1 GBXI01003042 JAD11250.1 ACPB03001633 GDIP01171335 JAJ52067.1 JRES01001517 KNC22305.1 CH379059 EAL34238.3 CH479180 EDW28377.1 GFDL01003480 JAV31565.1 GFWV01018996 MAA43724.1 GAKP01001649 JAC57303.1 GFTR01006733 JAW09693.1 OUUW01000006 SPP81622.1 CH933807 EDW13539.1 GECL01002607 JAP03517.1 RCHS01002807 RMX45643.1 CP012523 ALC39185.1 CM000157 EDW88790.1 CM000361 CM002910 EDX04501.1 KMY89484.1 AE014134 BT031279 AAF52908.1 ABY20520.1 AGB92856.1 CH480818 EDW52400.1 CH940649 EDW64548.1 GDHF01031981 JAI20333.1 CH954177 EDV58505.1 GANP01001228 JAB83240.1 GEFM01006610 JAP69186.1 CH902620 EDV32308.1 GACK01006752 JAA58282.1 GBBI01002768 JAC15944.1 GDRN01110638 JAI56875.1 CH916372 EDV98878.1 GFPF01013052 MAA24198.1 GEDV01004060 JAP84497.1 KA646459 AFP61088.1 CCAG010004816 EZ423570 ADD19846.1
KQ461073 KPJ09569.1 KQ971354 EFA06193.1 GEDC01000107 JAS37191.1 GEZM01022944 JAV88645.1 GL447678 EFN86194.1 GECZ01009337 JAS60432.1 GEDC01025861 JAS11437.1 KQ414699 KOC63450.1 KZ288386 PBC26480.1 KB632319 ERL92273.1 KQ434846 KZC08320.1 APGK01045645 BT128633 KB741037 AEE63590.1 ENN74687.1 NEVH01015306 PNF27077.1 GL768066 EFZ12593.1 GL888258 EGI63735.1 KQ435724 KOX78344.1 QOIP01000010 RLU17451.1 PNF27078.1 KQ981829 KYN35072.1 GBYB01012064 JAG81831.1 KQ980541 KYN15590.1 KQ976717 KYM76785.1 KQ983140 KYQ47074.1 NNAY01000510 OXU27924.1 GDIQ01074652 LRGB01000024 JAN20085.1 KZS21373.1 GL732583 EFX74497.1 KK107348 EZA52154.1 GDIP01033149 JAM70566.1 GDIQ01009631 JAN85106.1 IAAA01003043 IAAA01003044 LAA02825.1 AXCN02000845 GEBQ01010831 JAT29146.1 GANO01000714 JAB59157.1 ABLF02040831 GGFJ01004471 MBW53612.1 GFXV01001265 MBW13070.1 KK115530 KFM65368.1 AAAB01008835 JH431644 GGFK01005471 MBW38792.1 GAMD01002613 JAA98977.1 GGMR01012650 MBY25269.1 EAA05748.4 GGFL01004114 MBW68292.1 JR052538 AEY61718.1 DS235430 EEB15644.1 GDKW01001283 JAI55312.1 GGLE01005574 MBY09700.1 NCKU01003779 NCKU01002879 NCKU01002874 RWS06929.1 RWS08623.1 RWS08633.1 KB199650 ESP05184.1 UFQS01000111 UFQT01000111 SSW99799.1 SSX20179.1 GAMC01000905 JAC05651.1 GBGD01001239 JAC87650.1 GBXI01003042 JAD11250.1 ACPB03001633 GDIP01171335 JAJ52067.1 JRES01001517 KNC22305.1 CH379059 EAL34238.3 CH479180 EDW28377.1 GFDL01003480 JAV31565.1 GFWV01018996 MAA43724.1 GAKP01001649 JAC57303.1 GFTR01006733 JAW09693.1 OUUW01000006 SPP81622.1 CH933807 EDW13539.1 GECL01002607 JAP03517.1 RCHS01002807 RMX45643.1 CP012523 ALC39185.1 CM000157 EDW88790.1 CM000361 CM002910 EDX04501.1 KMY89484.1 AE014134 BT031279 AAF52908.1 ABY20520.1 AGB92856.1 CH480818 EDW52400.1 CH940649 EDW64548.1 GDHF01031981 JAI20333.1 CH954177 EDV58505.1 GANP01001228 JAB83240.1 GEFM01006610 JAP69186.1 CH902620 EDV32308.1 GACK01006752 JAA58282.1 GBBI01002768 JAC15944.1 GDRN01110638 JAI56875.1 CH916372 EDV98878.1 GFPF01013052 MAA24198.1 GEDV01004060 JAP84497.1 KA646459 AFP61088.1 CCAG010004816 EZ423570 ADD19846.1
Proteomes
UP000053268
UP000005204
UP000053240
UP000007266
UP000008237
UP000005203
+ More
UP000053825 UP000192223 UP000242457 UP000030742 UP000076502 UP000019118 UP000235965 UP000007755 UP000053105 UP000279307 UP000078541 UP000078492 UP000078540 UP000075809 UP000215335 UP000002358 UP000076858 UP000000305 UP000053097 UP000075903 UP000075886 UP000007819 UP000054359 UP000007062 UP000009046 UP000285301 UP000030746 UP000015103 UP000037069 UP000001819 UP000008744 UP000079169 UP000268350 UP000009192 UP000275408 UP000092553 UP000002282 UP000000304 UP000000803 UP000001292 UP000008792 UP000008711 UP000095300 UP000007801 UP000192221 UP000001070 UP000095301 UP000092445 UP000078200 UP000092444
UP000053825 UP000192223 UP000242457 UP000030742 UP000076502 UP000019118 UP000235965 UP000007755 UP000053105 UP000279307 UP000078541 UP000078492 UP000078540 UP000075809 UP000215335 UP000002358 UP000076858 UP000000305 UP000053097 UP000075903 UP000075886 UP000007819 UP000054359 UP000007062 UP000009046 UP000285301 UP000030746 UP000015103 UP000037069 UP000001819 UP000008744 UP000079169 UP000268350 UP000009192 UP000275408 UP000092553 UP000002282 UP000000304 UP000000803 UP000001292 UP000008792 UP000008711 UP000095300 UP000007801 UP000192221 UP000001070 UP000095301 UP000092445 UP000078200 UP000092444
Interpro
ProteinModelPortal
A0A194QGF7
A0A2H1WNJ8
H9IZS4
A0A194QW90
D6WPP3
A0A1B6EH15
+ More
A0A1Y1MW02 E2BDN2 A0A1B6GDA5 A0A1B6CDA9 A0A087ZQN5 A0A0L7QXV5 A0A1W4X482 A0A2A3E461 U4UIN8 A0A154PAW0 J3JZF1 A0A2J7QET0 E9J3G3 F4WPY4 A0A0N0U6N3 A0A3L8DAW3 A0A2J7QER1 A0A151JU35 A0A0C9R6V4 A0A151J194 A0A151HYU3 A0A151WGX3 A0A232FAQ5 K7IZG1 A0A0N8E077 E9H1J0 A0A026WAW6 A0A0P6AB63 A0A0P6IKD3 A0A182VAE0 A0A2L2Y6Q4 A0A182Q9F0 A0A1B6LZN6 U5EZI9 J9JTF1 A0A2M4BKR3 A0A1S4FQW4 A0A2H8TGA1 A0A087TJS9 A0A1S4H1L1 T1IX76 A0A2M4ADJ1 T1E8N2 A0A2S2P739 Q7QGE8 A0A2M4CSG8 V9IKQ5 E0VQI8 A0A0N7Z981 A0A2R5LJH8 A0A3S3RZB5 V4ALX1 A0A336K563 W8BWE7 A0A069DUM5 A0A0A1XK47 T1IFE6 A0A0P5CEX1 A0A0L0BQE2 Q29NI2 B4G7H7 A0A1Q3FVI4 A0A1S4EF74 A0A293MSB2 A0A034WU48 A0A224XNG6 A0A3B0JMS0 B4KJF9 A0A0V0G683 A0A3M6TW59 A0A0M4EC75 B4NZ95 B4Q8T6 Q9VKZ8 B4HWK5 B4LR09 A0A0K8U1E3 B3N970 V5HHU1 A0A1I8PCJ9 A0A131XU62 B3MPR0 L7M340 A0A1W4UUS3 A0A023F4P6 A0A0P4VVM3 B4JPP8 A0A1I8MRJ9 A0A1B0A536 A0A224Z2W2 A0A131YZR6 T1PCQ4 A0A1A9V9I6 D3TQ68
A0A1Y1MW02 E2BDN2 A0A1B6GDA5 A0A1B6CDA9 A0A087ZQN5 A0A0L7QXV5 A0A1W4X482 A0A2A3E461 U4UIN8 A0A154PAW0 J3JZF1 A0A2J7QET0 E9J3G3 F4WPY4 A0A0N0U6N3 A0A3L8DAW3 A0A2J7QER1 A0A151JU35 A0A0C9R6V4 A0A151J194 A0A151HYU3 A0A151WGX3 A0A232FAQ5 K7IZG1 A0A0N8E077 E9H1J0 A0A026WAW6 A0A0P6AB63 A0A0P6IKD3 A0A182VAE0 A0A2L2Y6Q4 A0A182Q9F0 A0A1B6LZN6 U5EZI9 J9JTF1 A0A2M4BKR3 A0A1S4FQW4 A0A2H8TGA1 A0A087TJS9 A0A1S4H1L1 T1IX76 A0A2M4ADJ1 T1E8N2 A0A2S2P739 Q7QGE8 A0A2M4CSG8 V9IKQ5 E0VQI8 A0A0N7Z981 A0A2R5LJH8 A0A3S3RZB5 V4ALX1 A0A336K563 W8BWE7 A0A069DUM5 A0A0A1XK47 T1IFE6 A0A0P5CEX1 A0A0L0BQE2 Q29NI2 B4G7H7 A0A1Q3FVI4 A0A1S4EF74 A0A293MSB2 A0A034WU48 A0A224XNG6 A0A3B0JMS0 B4KJF9 A0A0V0G683 A0A3M6TW59 A0A0M4EC75 B4NZ95 B4Q8T6 Q9VKZ8 B4HWK5 B4LR09 A0A0K8U1E3 B3N970 V5HHU1 A0A1I8PCJ9 A0A131XU62 B3MPR0 L7M340 A0A1W4UUS3 A0A023F4P6 A0A0P4VVM3 B4JPP8 A0A1I8MRJ9 A0A1B0A536 A0A224Z2W2 A0A131YZR6 T1PCQ4 A0A1A9V9I6 D3TQ68
PDB
5GJQ
E-value=7.10223e-128,
Score=1172
Ontologies
GO
Topology
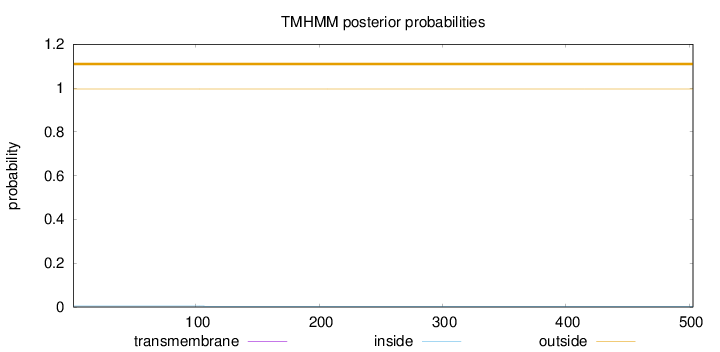
Length:
503
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00237
Exp number, first 60 AAs:
3e-05
Total prob of N-in:
0.00401
outside
1 - 503
Population Genetic Test Statistics
Pi
185.281627
Theta
148.46656
Tajima's D
-1.000394
CLR
1096.013158
CSRT
0.138243087845608
Interpretation
Uncertain
