Gene
KWMTBOMO11270 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012651
Annotation
PREDICTED:_uncharacterized_protein_LOC106139693_isoform_X1_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.186 Mitochondrial Reliability : 1.347
Sequence
CDS
ATGTCTTTGAAGAATAGGTTAATTGCATGTCTGCAAAAACTGAAGGAATACGATTACACTGACGTAGACAAGGCAAGAGCTAGTGACGCGAAGGCCGAGAATGCCCTCAACTTAATATACCAGACTGCTTTAAAGAAAACAACTTTTACACCAGAAGAGAAGAAAATTGTAGGTTCTCTTTTATTTGATGCCATCACACCGATACAAAGCAACATAGAAGGAACGCTCTGCGAATACAGAACTAGATTAGATACCTCTGTGCTTATGAAAAGATCTGCAATACAATTCTTGATTGACGAATACGGGAAGTTTCCAGTTGAGAATTCTATCTTGGCTACAAAGTTAGAGGAAGCCGGCATAGCAGAGAGCGTTGAAATACTCAATGAAATTATAAATAGGACATCGGCGGTCCTAGCAGGAGGAGCAGTTGGATGCCTCGCAGGACCGGCAGGAGCTGCATTGGGAGGCGTATACGGAGGCATGATTACAGACGGCTTGGTTTCTGCGGCTAAGAACGAGCCTGTAGGTATTTTTGAGTCAAGTGCAAATATAGGTAGAGACATTGCTTCAGGCAAAAATCCAATGGCTTCTGTGGGTTCCGCAAGTTTCAAAATCGCCATGGACGGTATAGGTGGCTTTGGCATGGGAGGAGTTTCTAGCGTTGCCAGTAAAGTCCCGGCGAAGGCCGTCGCAAAAACCATAGAAGGCACTGTAAAAAAAGCTGCTGCAACTATTGGGGAGGAGATGATTGAGAAGTCAGTGCAATTAACTGCAGAAGAAATGACAAAAATTGTGGCAAAGAAAGTGTCAGAAAAAGCCGTGAAAAAAGCTGTCGAAACTGCAGCAATACAAACACAAATGCTTCTGGTGTCTGCTCACGTCGGAGTAAATGCTAGTGAAAAAACCAAAGCTGAAATGCGTGCTGAAGAAGAACGTAAACAACAGAAACAAAAAGAAGAAGAACGTAAACAACAGAAACAAAAAGAAGAAGAAAAAAAGGCGCGTGAAAAGCGCGATAAAAAAAACACTGGATCGTGGCAACCGCCACGCGAGAACAGAAACAATAAAGAGTCCAATGTTGATAATGAAGAGTCCGACGAGGAGGTATTTGAAAAGTTCATGAAATTACTTAAACAGCTCTTGAAGCTTCTCTACGGCAATAACAGCGAACCGTTCACAAGAATTTTCGAGAATTTATCACCAGGCCAAATAGCGTATTTGGTGAAAATAATCAGAAACCTTAACTCAACTGGAATTTTACAGATAGTGTTAGATTTTACATTAAAACTGTTTAAATTGTACTTTTCTAAAGATATCTCATTCACAAATATCCTGAGTATGTTAGAATTCGTCGCAACTTACATTGAAGACCAGTTGTATTTTATGACCAAAACCAAAGGTAAACCGACTCGTCAATCAGATGCCGACCGGAATGAGCGTCGAAGGAACGCCATCAGAGAACTTCAAAACAAAACGGATATAATTCAGAAAATTGCTGATCTATTCAGACATTTACAAGCGGAAATCATAGCTGAAACGAGCGCGCCTAATATAATTTCCCCCTATTTCCGTGATATATTCTCAGCTGTGTATCATTATTACAAACACAGACGCGTGCCCGTGCCAGGCCATGAGAAGCTGACAGTCAAACAATATTTCGACTTGATAAGGATGGTAATTATAAAAATGAAAGAATCACCGGAATACAAAAAAATGCAGAAGGCCAGAAATGGAACGACCCTCACTTGTGAAATGTACGTGGTCTTGTTCGGTAGGATGTTCAGAATTATTATTACTGTTCGAGATGGTGAAATTGTATTGAGTACCGCCTATGTAGTCAAGCGTATGAACTTTAATGCCAAAGAAGGCATTCTTTACATAAAATTCGGTGATGGAGATGTACAGAAGTAA
Protein
MSLKNRLIACLQKLKEYDYTDVDKARASDAKAENALNLIYQTALKKTTFTPEEKKIVGSLLFDAITPIQSNIEGTLCEYRTRLDTSVLMKRSAIQFLIDEYGKFPVENSILATKLEEAGIAESVEILNEIINRTSAVLAGGAVGCLAGPAGAALGGVYGGMITDGLVSAAKNEPVGIFESSANIGRDIASGKNPMASVGSASFKIAMDGIGGFGMGGVSSVASKVPAKAVAKTIEGTVKKAAATIGEEMIEKSVQLTAEEMTKIVAKKVSEKAVKKAVETAAIQTQMLLVSAHVGVNASEKTKAEMRAEEERKQQKQKEEERKQQKQKEEEKKAREKRDKKNTGSWQPPRENRNNKESNVDNEESDEEVFEKFMKLLKQLLKLLYGNNSEPFTRIFENLSPGQIAYLVKIIRNLNSTGILQIVLDFTLKLFKLYFSKDISFTNILSMLEFVATYIEDQLYFMTKTKGKPTRQSDADRNERRRNAIRELQNKTDIIQKIADLFRHLQAEIIAETSAPNIISPYFRDIFSAVYHYYKHRRVPVPGHEKLTVKQYFDLIRMVIIKMKESPEYKKMQKARNGTTLTCEMYVVLFGRMFRIIITVRDGEIVLSTAYVVKRMNFNAKEGILYIKFGDGDVQK
Summary
Uniprot
ProteinModelPortal
Ontologies
GO
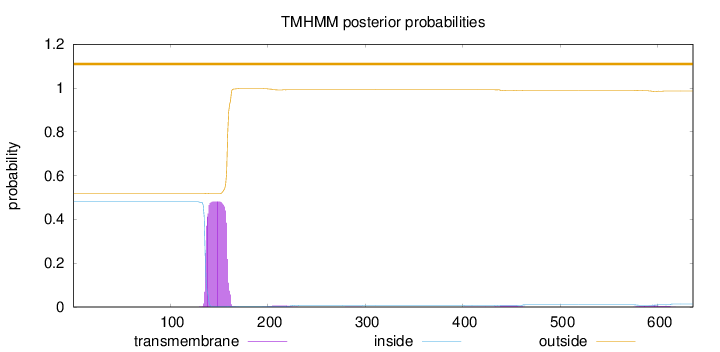
Topology
Length:
636
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
11.42831
Exp number, first 60 AAs:
0.00024
Total prob of N-in:
0.48027
outside
1 - 636
Population Genetic Test Statistics
Pi
273.728122
Theta
173.686184
Tajima's D
0.559429
CLR
0.765212
CSRT
0.531923403829809
Interpretation
Uncertain
