Pre Gene Modal
BGIBMGA012582
Annotation
PREDICTED:_cleavage_and_polyadenylation_specificity_factor_73_[Bombyx_mori]
Full name
Ubiquitinyl hydrolase 1
Location in the cell
Cytoplasmic Reliability : 1.225 PlasmaMembrane Reliability : 1.739
Sequence
CDS
ATGACTACAAATCCAAGAAAAGGCACTGATCCAGTGCCTGCGGAGGAAAGCGATCAATTGACAATAAGACCACTTGGAGCTGGCCAAGAAGTTGGCAGATCATGCATCATGTTGGAGTTCAAAGGGAAAAAGATAATGTTGGACTGTGGCATCCATCCTGGTTTGTCTGGAATGGACGCCCTACCCTTCATAGACCTTATAGAGGCAGACGAAGTGGATCTTCTGTTAGTTTCACATTTTCATTTGGACCACAGTGGAGCCCTTCCGTGGTTCCTGACGAAGACTTCCTTCAAAGGTCGAGTTTTCATGACACATGCCACTAAGGCTATCTACAGATGGCTCGTTTCTGATTATATTAAAGTTAGCAACATATCAACAGAACAAATGTTGTATACGGAATCGGATCTAGAAAATTCAATGGACAGAATAGAAACTATAAACTTCCATGAAGAAAGAGATGTACGGGGTGTCAAATTTTGGGCTTACAATGCTGGACATGTACTCGGAGCAGCCATGTTTATGATTGAAATAGCTGGAGTCAAGATATTATACACGGGAGATTTTTCAAGACAAGAGGATAGACATTTGATGGCTGCAGAGATACCGACGGTTCATCCTGATGTTTTGATTACTGAGTCCACGTACGGCACGCACATCCACGAGAAGCGCGAGGAGCGGGAGAGTCGCTTCACGGGGCTGGTCAGCGACATCGTGGGGCGCGGCGGCCGCTGCCTCATCCCCGTGTTCGCGCTGGGCCGCGCGCAGGAGCTGCTGCTCATACTCGACGAGTACTGGTCGCTGCACCCCGAGCTGCAAGACATTCCCATATACTACGCGTCCTCACTGGCCAAGAAGTGCATGGCGGTGTACCAGACCTATGTTAACGCGATGAACGACCGCATCCGGCGCCAGATCGCTATCAACAACCCCTTCGTGTTTAGACACATCTCCAACCTGAAGTTGGGCACAAATGACTTTAACTCTGTCTAA
Protein
MTTNPRKGTDPVPAEESDQLTIRPLGAGQEVGRSCIMLEFKGKKIMLDCGIHPGLSGMDALPFIDLIEADEVDLLLVSHFHLDHSGALPWFLTKTSFKGRVFMTHATKAIYRWLVSDYIKVSNISTEQMLYTESDLENSMDRIETINFHEERDVRGVKFWAYNAGHVLGAAMFMIEIAGVKILYTGDFSRQEDRHLMAAEIPTVHPDVLITESTYGTHIHEKREERESRFTGLVSDIVGRGGRCLIPVFALGRAQELLLILDEYWSLHPELQDIPIYYASSLAKKCMAVYQTYVNAMNDRIRRQIAINNPFVFRHISNLKLGTNDFNSV
Summary
Catalytic Activity
Thiol-dependent hydrolysis of ester, thioester, amide, peptide and isopeptide bonds formed by the C-terminal Gly of ubiquitin (a 76-residue protein attached to proteins as an intracellular targeting signal).
Similarity
Belongs to the peptidase C19 family.
Uniprot
A0A2H1VQI7
A0A2A4JX46
A0A0N1IMS2
A0A0N1IEX9
A0A212F2I3
A0A2J7R5C5
+ More
A0A067RDR8 A0A139WFW0 A0A1Y1LBJ4 A0A1B6DJS3 E2AIH9 A0A1B6HGW1 A0A1B6ET64 A0A1B6KSK1 A0A2A3EHP0 A0A088A3T2 A0A154PS55 A0A0L7QWD3 A0A1B6I2X2 A0A026WY67 K7J2M9 A0A232F768 A0A0J7KBN9 E9J3A8 A0A195B7C4 A0A195FNY7 A0A158NGY1 A0A151ID90 A0A195DWF4 F4X126 T1JIA8 E2BWA8 B0X8N8 A0A1Q3F4J9 A0A0P5UYR6 A0A0P5B7F1 A0A0P5UET1 E9H6R0 A0A0P5ZCM9 A0A0P5VMA9 A0A0P4Y968 A0A0P5MRA6 A0A0P5LUC8 A0A0P5HI95 A0A0P5RC32 A0A0P6G968 Q170J6 L7LUX8 A0A224YLX6 A0A131Z7V5 A0A151XD31 A0A0P6F1X4 A0A0P5VCY2 A0A336MGA6 A0A336M7S8 A0A1B0GJR3 A0A0A9YYP8 A0A1L8DH05 A0A023F454 T1HZV1 A0A069DVS5 A0A0V0G9S7 A0A0P5V502 B7P2D2 A0A1L8DGC9 V5H4V8 A0A0L0CNN5 A0A2R7X216 A0A1I8PMU4 A0A2R5LL73 A0A1A9W2T5 A0A1A9Y9E4 A0A1A9ZEA1 A0A1I8MBA9 A0A1B0BGV6 E0VDY7 W8CAQ0 A0A0K8VB67 A0A1A9V5Z7 A0A0A1WZH2 A0A034VDT8 A0A084W029 A0A131XG05 A0A182IZ35 A0A1W4UK58 A0A182LYH6 A0A1J1IAV5 A0A182K254 B4M3X4 A0A0P6BGC3 A0A182VW06 B4K8L8 A0A182X9D1 Q299N6 B4G5H0 A0A182YHX5 A0A182L8K4 A0A182TS82 A0A182HKB7 A0A182RSD2
A0A067RDR8 A0A139WFW0 A0A1Y1LBJ4 A0A1B6DJS3 E2AIH9 A0A1B6HGW1 A0A1B6ET64 A0A1B6KSK1 A0A2A3EHP0 A0A088A3T2 A0A154PS55 A0A0L7QWD3 A0A1B6I2X2 A0A026WY67 K7J2M9 A0A232F768 A0A0J7KBN9 E9J3A8 A0A195B7C4 A0A195FNY7 A0A158NGY1 A0A151ID90 A0A195DWF4 F4X126 T1JIA8 E2BWA8 B0X8N8 A0A1Q3F4J9 A0A0P5UYR6 A0A0P5B7F1 A0A0P5UET1 E9H6R0 A0A0P5ZCM9 A0A0P5VMA9 A0A0P4Y968 A0A0P5MRA6 A0A0P5LUC8 A0A0P5HI95 A0A0P5RC32 A0A0P6G968 Q170J6 L7LUX8 A0A224YLX6 A0A131Z7V5 A0A151XD31 A0A0P6F1X4 A0A0P5VCY2 A0A336MGA6 A0A336M7S8 A0A1B0GJR3 A0A0A9YYP8 A0A1L8DH05 A0A023F454 T1HZV1 A0A069DVS5 A0A0V0G9S7 A0A0P5V502 B7P2D2 A0A1L8DGC9 V5H4V8 A0A0L0CNN5 A0A2R7X216 A0A1I8PMU4 A0A2R5LL73 A0A1A9W2T5 A0A1A9Y9E4 A0A1A9ZEA1 A0A1I8MBA9 A0A1B0BGV6 E0VDY7 W8CAQ0 A0A0K8VB67 A0A1A9V5Z7 A0A0A1WZH2 A0A034VDT8 A0A084W029 A0A131XG05 A0A182IZ35 A0A1W4UK58 A0A182LYH6 A0A1J1IAV5 A0A182K254 B4M3X4 A0A0P6BGC3 A0A182VW06 B4K8L8 A0A182X9D1 Q299N6 B4G5H0 A0A182YHX5 A0A182L8K4 A0A182TS82 A0A182HKB7 A0A182RSD2
EC Number
3.4.19.12
Pubmed
26354079
22118469
24845553
18362917
19820115
28004739
+ More
20798317 24508170 30249741 20075255 28648823 21282665 21347285 21719571 21292972 17510324 25576852 28797301 26830274 25401762 26823975 25474469 26334808 25765539 26108605 25315136 20566863 24495485 25830018 25348373 24438588 28049606 17994087 15632085 25244985 20966253
20798317 24508170 30249741 20075255 28648823 21282665 21347285 21719571 21292972 17510324 25576852 28797301 26830274 25401762 26823975 25474469 26334808 25765539 26108605 25315136 20566863 24495485 25830018 25348373 24438588 28049606 17994087 15632085 25244985 20966253
EMBL
ODYU01003846
SOQ43113.1
NWSH01000411
PCG76587.1
KQ459449
KPJ00887.1
+ More
KQ461184 KPJ07521.1 AGBW02010741 OWR47933.1 NEVH01006994 PNF36035.1 KK852527 KDR22016.1 KQ971352 KYB26687.1 GEZM01063106 JAV69305.1 GEDC01011420 JAS25878.1 GL439791 EFN66769.1 GECU01033749 JAS73957.1 GECZ01028698 GECZ01022214 JAS41071.1 JAS47555.1 GEBQ01025544 JAT14433.1 KZ288239 PBC31295.1 KQ435041 KZC14174.1 KQ414714 KOC62927.1 GECU01026432 JAS81274.1 KK107063 QOIP01000005 EZA60982.1 RLU23084.1 NNAY01000831 OXU26288.1 LBMM01010055 KMQ87697.1 GL768061 EFZ12664.1 KQ976574 KYM80155.1 KQ981387 KYN42027.1 ADTU01015321 KQ977985 KYM98347.1 KQ980204 KYN17240.1 GL888525 EGI59837.1 JH431327 GL451103 EFN80012.1 DS232495 EDS42630.1 GFDL01012592 JAV22453.1 GDIP01107537 JAL96177.1 GDIP01189028 JAJ34374.1 GDIP01115436 JAL88278.1 GL732598 EFX72593.1 GDIP01045707 GDIQ01036911 LRGB01002993 JAM58008.1 JAN57826.1 KZS05118.1 GDIP01098137 JAM05578.1 GDIP01234527 JAI88874.1 GDIQ01175248 JAK76477.1 GDIQ01175249 JAK76476.1 GDIQ01227098 JAK24627.1 GDIQ01102840 JAL48886.1 GDIQ01055463 JAN39274.1 CH477473 EAT40345.1 GACK01009622 JAA55412.1 GFPF01006809 MAA17955.1 GEDV01002306 JAP86251.1 KQ982294 KYQ58286.1 GDIQ01055462 JAN39275.1 GDIP01102041 JAM01674.1 UFQS01000883 UFQT01000883 SSX07493.1 SSX27833.1 UFQS01000673 UFQT01000673 SSX06015.1 SSX26372.1 AJWK01022144 AJWK01022145 AJWK01022146 GBHO01007381 GBHO01007380 GBRD01013468 GDHC01004914 JAG36223.1 JAG36224.1 JAG52358.1 JAQ13715.1 GFDF01008345 JAV05739.1 GBBI01002432 JAC16280.1 ACPB03022312 GBGD01000691 JAC88198.1 GECL01001986 JAP04138.1 GDIP01104938 JAL98776.1 ABJB010540581 DS621961 EEC00754.1 GFDF01008566 JAV05518.1 GANP01006423 JAB78045.1 JRES01000142 KNC33866.1 KK856473 PTY25849.1 GGLE01006137 MBY10263.1 JXJN01014010 DS235088 EEB11593.1 GAMC01005546 JAC01010.1 GDHF01016201 JAI36113.1 GBXI01010226 JAD04066.1 GAKP01018690 GAKP01018688 JAC40262.1 ATLV01019081 KE525262 KFB43573.1 GEFH01003074 JAP65507.1 AXCM01001239 CVRI01000044 CRK96692.1 CH940652 EDW59335.1 GDIP01015566 JAM88149.1 CH933806 EDW14417.1 CM000070 EAL27667.2 CH479179 EDW24836.1 APCN01000825
KQ461184 KPJ07521.1 AGBW02010741 OWR47933.1 NEVH01006994 PNF36035.1 KK852527 KDR22016.1 KQ971352 KYB26687.1 GEZM01063106 JAV69305.1 GEDC01011420 JAS25878.1 GL439791 EFN66769.1 GECU01033749 JAS73957.1 GECZ01028698 GECZ01022214 JAS41071.1 JAS47555.1 GEBQ01025544 JAT14433.1 KZ288239 PBC31295.1 KQ435041 KZC14174.1 KQ414714 KOC62927.1 GECU01026432 JAS81274.1 KK107063 QOIP01000005 EZA60982.1 RLU23084.1 NNAY01000831 OXU26288.1 LBMM01010055 KMQ87697.1 GL768061 EFZ12664.1 KQ976574 KYM80155.1 KQ981387 KYN42027.1 ADTU01015321 KQ977985 KYM98347.1 KQ980204 KYN17240.1 GL888525 EGI59837.1 JH431327 GL451103 EFN80012.1 DS232495 EDS42630.1 GFDL01012592 JAV22453.1 GDIP01107537 JAL96177.1 GDIP01189028 JAJ34374.1 GDIP01115436 JAL88278.1 GL732598 EFX72593.1 GDIP01045707 GDIQ01036911 LRGB01002993 JAM58008.1 JAN57826.1 KZS05118.1 GDIP01098137 JAM05578.1 GDIP01234527 JAI88874.1 GDIQ01175248 JAK76477.1 GDIQ01175249 JAK76476.1 GDIQ01227098 JAK24627.1 GDIQ01102840 JAL48886.1 GDIQ01055463 JAN39274.1 CH477473 EAT40345.1 GACK01009622 JAA55412.1 GFPF01006809 MAA17955.1 GEDV01002306 JAP86251.1 KQ982294 KYQ58286.1 GDIQ01055462 JAN39275.1 GDIP01102041 JAM01674.1 UFQS01000883 UFQT01000883 SSX07493.1 SSX27833.1 UFQS01000673 UFQT01000673 SSX06015.1 SSX26372.1 AJWK01022144 AJWK01022145 AJWK01022146 GBHO01007381 GBHO01007380 GBRD01013468 GDHC01004914 JAG36223.1 JAG36224.1 JAG52358.1 JAQ13715.1 GFDF01008345 JAV05739.1 GBBI01002432 JAC16280.1 ACPB03022312 GBGD01000691 JAC88198.1 GECL01001986 JAP04138.1 GDIP01104938 JAL98776.1 ABJB010540581 DS621961 EEC00754.1 GFDF01008566 JAV05518.1 GANP01006423 JAB78045.1 JRES01000142 KNC33866.1 KK856473 PTY25849.1 GGLE01006137 MBY10263.1 JXJN01014010 DS235088 EEB11593.1 GAMC01005546 JAC01010.1 GDHF01016201 JAI36113.1 GBXI01010226 JAD04066.1 GAKP01018690 GAKP01018688 JAC40262.1 ATLV01019081 KE525262 KFB43573.1 GEFH01003074 JAP65507.1 AXCM01001239 CVRI01000044 CRK96692.1 CH940652 EDW59335.1 GDIP01015566 JAM88149.1 CH933806 EDW14417.1 CM000070 EAL27667.2 CH479179 EDW24836.1 APCN01000825
Proteomes
UP000218220
UP000053268
UP000053240
UP000007151
UP000235965
UP000027135
+ More
UP000007266 UP000000311 UP000242457 UP000005203 UP000076502 UP000053825 UP000053097 UP000279307 UP000002358 UP000215335 UP000036403 UP000078540 UP000078541 UP000005205 UP000078542 UP000078492 UP000007755 UP000008237 UP000002320 UP000000305 UP000076858 UP000008820 UP000075809 UP000092461 UP000015103 UP000001555 UP000037069 UP000095300 UP000091820 UP000092443 UP000092445 UP000095301 UP000092460 UP000009046 UP000078200 UP000030765 UP000075880 UP000192221 UP000075883 UP000183832 UP000075881 UP000008792 UP000075920 UP000009192 UP000076407 UP000001819 UP000008744 UP000076408 UP000075882 UP000075902 UP000075840 UP000075900
UP000007266 UP000000311 UP000242457 UP000005203 UP000076502 UP000053825 UP000053097 UP000279307 UP000002358 UP000215335 UP000036403 UP000078540 UP000078541 UP000005205 UP000078542 UP000078492 UP000007755 UP000008237 UP000002320 UP000000305 UP000076858 UP000008820 UP000075809 UP000092461 UP000015103 UP000001555 UP000037069 UP000095300 UP000091820 UP000092443 UP000092445 UP000095301 UP000092460 UP000009046 UP000078200 UP000030765 UP000075880 UP000192221 UP000075883 UP000183832 UP000075881 UP000008792 UP000075920 UP000009192 UP000076407 UP000001819 UP000008744 UP000076408 UP000075882 UP000075902 UP000075840 UP000075900
Pfam
Interpro
IPR011108
RMMBL
+ More
IPR036866 RibonucZ/Hydroxyglut_hydro
IPR022712 Beta_Casp
IPR021718 CPSF73-100_C
IPR001279 Metallo-B-lactamas
IPR013087 Znf_C2H2_type
IPR020846 MFS_dom
IPR011701 MFS
IPR036259 MFS_trans_sf
IPR041432 UBP13_Znf-UBP_var
IPR018200 USP_CS
IPR038765 Papain-like_cys_pep_sf
IPR015940 UBA
IPR001394 Peptidase_C19_UCH
IPR001607 Znf_UBP
IPR013083 Znf_RING/FYVE/PHD
IPR028889 USP_dom
IPR036866 RibonucZ/Hydroxyglut_hydro
IPR022712 Beta_Casp
IPR021718 CPSF73-100_C
IPR001279 Metallo-B-lactamas
IPR013087 Znf_C2H2_type
IPR020846 MFS_dom
IPR011701 MFS
IPR036259 MFS_trans_sf
IPR041432 UBP13_Znf-UBP_var
IPR018200 USP_CS
IPR038765 Papain-like_cys_pep_sf
IPR015940 UBA
IPR001394 Peptidase_C19_UCH
IPR001607 Znf_UBP
IPR013083 Znf_RING/FYVE/PHD
IPR028889 USP_dom
Gene 3D
CDD
ProteinModelPortal
A0A2H1VQI7
A0A2A4JX46
A0A0N1IMS2
A0A0N1IEX9
A0A212F2I3
A0A2J7R5C5
+ More
A0A067RDR8 A0A139WFW0 A0A1Y1LBJ4 A0A1B6DJS3 E2AIH9 A0A1B6HGW1 A0A1B6ET64 A0A1B6KSK1 A0A2A3EHP0 A0A088A3T2 A0A154PS55 A0A0L7QWD3 A0A1B6I2X2 A0A026WY67 K7J2M9 A0A232F768 A0A0J7KBN9 E9J3A8 A0A195B7C4 A0A195FNY7 A0A158NGY1 A0A151ID90 A0A195DWF4 F4X126 T1JIA8 E2BWA8 B0X8N8 A0A1Q3F4J9 A0A0P5UYR6 A0A0P5B7F1 A0A0P5UET1 E9H6R0 A0A0P5ZCM9 A0A0P5VMA9 A0A0P4Y968 A0A0P5MRA6 A0A0P5LUC8 A0A0P5HI95 A0A0P5RC32 A0A0P6G968 Q170J6 L7LUX8 A0A224YLX6 A0A131Z7V5 A0A151XD31 A0A0P6F1X4 A0A0P5VCY2 A0A336MGA6 A0A336M7S8 A0A1B0GJR3 A0A0A9YYP8 A0A1L8DH05 A0A023F454 T1HZV1 A0A069DVS5 A0A0V0G9S7 A0A0P5V502 B7P2D2 A0A1L8DGC9 V5H4V8 A0A0L0CNN5 A0A2R7X216 A0A1I8PMU4 A0A2R5LL73 A0A1A9W2T5 A0A1A9Y9E4 A0A1A9ZEA1 A0A1I8MBA9 A0A1B0BGV6 E0VDY7 W8CAQ0 A0A0K8VB67 A0A1A9V5Z7 A0A0A1WZH2 A0A034VDT8 A0A084W029 A0A131XG05 A0A182IZ35 A0A1W4UK58 A0A182LYH6 A0A1J1IAV5 A0A182K254 B4M3X4 A0A0P6BGC3 A0A182VW06 B4K8L8 A0A182X9D1 Q299N6 B4G5H0 A0A182YHX5 A0A182L8K4 A0A182TS82 A0A182HKB7 A0A182RSD2
A0A067RDR8 A0A139WFW0 A0A1Y1LBJ4 A0A1B6DJS3 E2AIH9 A0A1B6HGW1 A0A1B6ET64 A0A1B6KSK1 A0A2A3EHP0 A0A088A3T2 A0A154PS55 A0A0L7QWD3 A0A1B6I2X2 A0A026WY67 K7J2M9 A0A232F768 A0A0J7KBN9 E9J3A8 A0A195B7C4 A0A195FNY7 A0A158NGY1 A0A151ID90 A0A195DWF4 F4X126 T1JIA8 E2BWA8 B0X8N8 A0A1Q3F4J9 A0A0P5UYR6 A0A0P5B7F1 A0A0P5UET1 E9H6R0 A0A0P5ZCM9 A0A0P5VMA9 A0A0P4Y968 A0A0P5MRA6 A0A0P5LUC8 A0A0P5HI95 A0A0P5RC32 A0A0P6G968 Q170J6 L7LUX8 A0A224YLX6 A0A131Z7V5 A0A151XD31 A0A0P6F1X4 A0A0P5VCY2 A0A336MGA6 A0A336M7S8 A0A1B0GJR3 A0A0A9YYP8 A0A1L8DH05 A0A023F454 T1HZV1 A0A069DVS5 A0A0V0G9S7 A0A0P5V502 B7P2D2 A0A1L8DGC9 V5H4V8 A0A0L0CNN5 A0A2R7X216 A0A1I8PMU4 A0A2R5LL73 A0A1A9W2T5 A0A1A9Y9E4 A0A1A9ZEA1 A0A1I8MBA9 A0A1B0BGV6 E0VDY7 W8CAQ0 A0A0K8VB67 A0A1A9V5Z7 A0A0A1WZH2 A0A034VDT8 A0A084W029 A0A131XG05 A0A182IZ35 A0A1W4UK58 A0A182LYH6 A0A1J1IAV5 A0A182K254 B4M3X4 A0A0P6BGC3 A0A182VW06 B4K8L8 A0A182X9D1 Q299N6 B4G5H0 A0A182YHX5 A0A182L8K4 A0A182TS82 A0A182HKB7 A0A182RSD2
PDB
2I7V
E-value=6.61516e-161,
Score=1455
Ontologies
GO
Topology
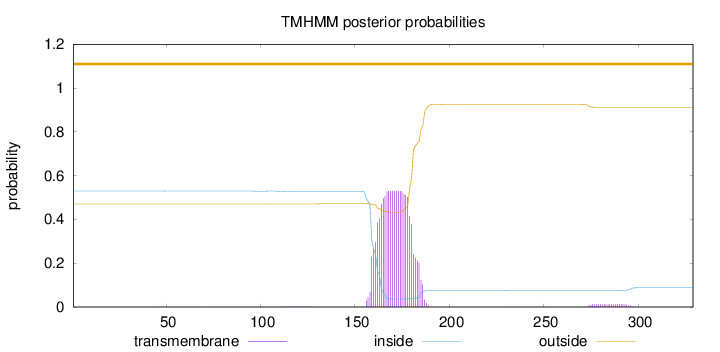
Length:
329
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
11.7882
Exp number, first 60 AAs:
0.00544
Total prob of N-in:
0.52977
outside
1 - 329
Population Genetic Test Statistics
Pi
227.966377
Theta
189.135586
Tajima's D
0.601723
CLR
0.133622
CSRT
0.542872856357182
Interpretation
Uncertain
