Gene
KWMTBOMO11252
Pre Gene Modal
BGIBMGA012624
Annotation
PREDICTED:_delta(24)-sterol_reductase-like_isoform_X1_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.515
Sequence
CDS
ATGTTAAGCAGACGAGCATACGGCCCAGCTGATGGTGAGTGTTTACTGTCGCTCATGGACGTCAGCAATGCCAGGGGCAGAGCCAAGCTGCTGCCTACCGTTAAGTACTCTCCACAAGCCTCGTTTGAAGAAGGACATGTCATAGCGCTCCGGAAACACCGCGAGGGTAGCTCATTCCACAGGATGATGGTGCATTTGAAAGCTAGACTGATTAGTTGGTTGGAGAACAACCGGGAGTTAGTAGTGGCGCTGTTCTGTCTGCCAGCCAGCTTCCTGTTCACGGTAGCGCTGCAGCTGCGAGCGCACTTACGGAGACTCGTCGCTCGCGCCGACCAGCACGAGAGGGCCGTTAAAGAGATCCAGGCTAGGGTCCAGGAATGGAACAAGCTTCCGTCACGAGACCGTCGACTGCTGTGCACGTCGAGACCCAACTGGTTGTCTCTCTCTACTACGTTTTTCGAAAAACATCTCCATCATCGAGTTCCTATACCGCTTTACGACATATTGGAGCTGGATGAACAAGCTATGACTGTCAGAGTCGAGCCAATGGTGACCATCGGCGACATCACAGAATACCTCATACCGAAGGGCTATTCATTGGCCGTCACTATTGAATTGGTTGACGCTACACTTGGAGGTCTTGCATTCGGCACTGGAATGTCCACTCACTCCCACAAAGCGGGCTTGTACCACGAGACTATTACTGCGTATGAAGTGGTTCTCGGTGACGGGTCCCTGGTCACAGCTACAGCTGACAATGAACACTCTGATCTCTTCAAAGCCCTCCCGTGGTCGCATGGTAGTCTTGGCTTCCTTGTGGCGCTCACGTTGAAAATTGTCAAAGTAAAACCATTCATTAAGATCAAGTACACTCCGGTTGAAGGACAGAAGGAATATTGCAACATGCTGCGAGAACTTTCTGGAACGTACGAAAGAGAACCTAAAGAATTCCCCGATTTTCTCGAGGCAACAATCTTTAGTGAGAACGAGGCGGTCGTCATGTCAGGAGACTACTGCGACCGCGACCCGACTGCGCCAGTGAATCACTGCTCGAGGTGGTACAAGCCGTGGTTTTACAAACACGTACAGTCCTTCTTGAAGAAGGGGGAGAGCGTCGAGTTGATACCGTTGAAAGATTATTTACTCCGACACAACAGAGCTATCTTCTGGGTCGTCGAGGACATGCTCTCATTCGGAAACGACGCCTTGTTCCGATACTTGTTCGGTTGGCTGCTGCCCCCCAAGCCCGCCTTTTTAAAATTCACGACGACGCCGGGGGTGCGAGCCTACACATTCACGAAGCAAGTCTTCCAAGACATAGTGTTGCCGATCAGTGAACTGGAACGGCAGATCGAGATGGCCACGCAATTGTTTGACAAATTCCCGCTGCTTGTGTACCCGTGTAAAGTGATCGATCGCGGTCCATCGTCCGGACAACTGAAACGACCCCACTCTAAGTACTTGGTCCCGGGAACAAACTATGCGATGTACAATGATTTGGGAGTTTACGGCGTCCCGGGGAAGGTCAAAGAGAAGAAGCCATACAGCCCGGTGACGGCTATGAGAAAGATGGAAGAGTTCACGAGAGAAGTGGGTGGTTTTTCATTTCTTTACGCCGATATATTCATGACCCGAGAGGAGTTTGAGGCCATGTTTGATTTATCTCTGTACGAGAGAGTTCGAAGGAAGTATGGAGCTGAAGGAGCCTTCCCACATTTATATGTTAAAGTCAAGCCAGAGATTGATGTTTTCGCTATTGGTGAAGAGAACGCTGTCCAGTGA
Protein
MLSRRAYGPADGECLLSLMDVSNARGRAKLLPTVKYSPQASFEEGHVIALRKHREGSSFHRMMVHLKARLISWLENNRELVVALFCLPASFLFTVALQLRAHLRRLVARADQHERAVKEIQARVQEWNKLPSRDRRLLCTSRPNWLSLSTTFFEKHLHHRVPIPLYDILELDEQAMTVRVEPMVTIGDITEYLIPKGYSLAVTIELVDATLGGLAFGTGMSTHSHKAGLYHETITAYEVVLGDGSLVTATADNEHSDLFKALPWSHGSLGFLVALTLKIVKVKPFIKIKYTPVEGQKEYCNMLRELSGTYEREPKEFPDFLEATIFSENEAVVMSGDYCDRDPTAPVNHCSRWYKPWFYKHVQSFLKKGESVELIPLKDYLLRHNRAIFWVVEDMLSFGNDALFRYLFGWLLPPKPAFLKFTTTPGVRAYTFTKQVFQDIVLPISELERQIEMATQLFDKFPLLVYPCKVIDRGPSSGQLKRPHSKYLVPGTNYAMYNDLGVYGVPGKVKEKKPYSPVTAMRKMEEFTREVGGFSFLYADIFMTREEFEAMFDLSLYERVRRKYGAEGAFPHLYVKVKPEIDVFAIGEENAVQ
Summary
Uniprot
H9JSW2
A0A2A4JYM3
A0A2H1VSB6
A0A2W1BPJ6
A0A0N1ID81
A0A0N1PGV7
+ More
A0A212F2H7 A0A0L7L9T6 D6WTX2 V5GXQ9 A0A1W4WBU7 A0A0P5KUL6 A0A0P5YW80 A0A0P5GEV7 A0A0P5J9A1 A0A0N8DB92 A0A0P5YK95 A0A1D2NDB4 C7DR07 A0A0P5WAM6 A0A0P6A5I7 A0A0P6BAP2 A0A0P5PHY4 A0A1B6JIJ3 A0A267FXT9 A0A226F1P3 A0A267H929 A0A267FY07 A0A1B6C9D4 A0A3R7T0B8 A0A0P5NP72 A0A0P5CP71 A0A1W2W6U8 A0A0P4WH00 E3MJD7 Q9XVZ2 A0A261CPQ7 F6ZXA7 H2ZIN9 A0A2H2I517 A0A1I7TRT2 G0MAQ7 A0A1I7SVI7 A0A2G5T0E7 A8X449 A0A0P5CH18 A0A0P4W8Z8 A0A0P6HFF3 H3EYA8 A0A2A2KPH2 A0A261BD58 A0A0P5BQE7 A0A2A2KPD6 A0A2A2KVH6 A0A0P5RHF8 A0A0P5XTY9 A0A0P5RPZ0 A0A1I2FH59 A0A2P4WHZ1 A0A2S9YWS8 A0A2S9YJN9 A0A0C2DFF7 A0A1G0GBE5 A0A367LEX4 A0A369H047 A0A094IY66 A0A179I8B4 A0A162MZF9 A0A369GVF9 W7TVX8 A0A137NTE9 A0A1Y1XUM5 A0A2H4S6G3 A0A1V4JZG3 A0A218UXF9 A0A3L8SQF0 K7F6B9 Q5ZIF2 G1K8B5 A0A3M0KCB9 A0A093Q065 M7B6K0 A0A1S9CYJ4 A0A0S8BZW2
A0A212F2H7 A0A0L7L9T6 D6WTX2 V5GXQ9 A0A1W4WBU7 A0A0P5KUL6 A0A0P5YW80 A0A0P5GEV7 A0A0P5J9A1 A0A0N8DB92 A0A0P5YK95 A0A1D2NDB4 C7DR07 A0A0P5WAM6 A0A0P6A5I7 A0A0P6BAP2 A0A0P5PHY4 A0A1B6JIJ3 A0A267FXT9 A0A226F1P3 A0A267H929 A0A267FY07 A0A1B6C9D4 A0A3R7T0B8 A0A0P5NP72 A0A0P5CP71 A0A1W2W6U8 A0A0P4WH00 E3MJD7 Q9XVZ2 A0A261CPQ7 F6ZXA7 H2ZIN9 A0A2H2I517 A0A1I7TRT2 G0MAQ7 A0A1I7SVI7 A0A2G5T0E7 A8X449 A0A0P5CH18 A0A0P4W8Z8 A0A0P6HFF3 H3EYA8 A0A2A2KPH2 A0A261BD58 A0A0P5BQE7 A0A2A2KPD6 A0A2A2KVH6 A0A0P5RHF8 A0A0P5XTY9 A0A0P5RPZ0 A0A1I2FH59 A0A2P4WHZ1 A0A2S9YWS8 A0A2S9YJN9 A0A0C2DFF7 A0A1G0GBE5 A0A367LEX4 A0A369H047 A0A094IY66 A0A179I8B4 A0A162MZF9 A0A369GVF9 W7TVX8 A0A137NTE9 A0A1Y1XUM5 A0A2H4S6G3 A0A1V4JZG3 A0A218UXF9 A0A3L8SQF0 K7F6B9 Q5ZIF2 G1K8B5 A0A3M0KCB9 A0A093Q065 M7B6K0 A0A1S9CYJ4 A0A0S8BZW2
Pubmed
EMBL
BABH01035994
NWSH01000411
PCG76593.1
ODYU01003846
SOQ43114.1
KZ149965
+ More
PZC76211.1 KQ461184 KPJ07522.1 KQ459449 KPJ00888.1 AGBW02010741 OWR47932.1 JTDY01002142 KOB72016.1 KQ971352 EFA07314.1 GALX01003323 JAB65143.1 GDIQ01184362 JAK67363.1 GDIP01052309 JAM51406.1 GDIQ01249136 JAK02589.1 GDIQ01205159 JAK46566.1 GDIP01050711 JAM53004.1 GDIP01070601 LRGB01002994 JAM33114.1 KZS05093.1 LJIJ01000081 ODN03240.1 GQ199603 GL732567 ACT20728.1 EFX76668.1 GDIP01088803 JAM14912.1 GDIP01046912 JAM56803.1 GDIP01019387 JAM84328.1 GDIQ01133836 JAL17890.1 GECU01008590 JAS99116.1 NIVC01000721 PAA77819.1 LNIX01000001 OXA63121.1 NIVC01000017 PAA94059.1 NIVC01000720 PAA77842.1 GEDC01027191 JAS10107.1 QCYY01000619 ROT83942.1 GDIQ01149832 JAL01894.1 GDIP01171442 JAJ51960.1 GDRN01062447 JAI65069.1 DS268450 EFP03687.1 BX284606 CAA22461.1 NIPN01000002 OZG23886.1 EAAA01002802 GL379788 EGT40606.1 PDUG01000006 PIC20596.1 HE601041 CAP27409.1 GDIP01171444 JAJ51958.1 GDRN01062450 JAI65068.1 GDIQ01019818 JAN74919.1 LIAE01008038 PAV75789.1 NMWX01000001 OZG07670.1 GDIP01181780 JAJ41622.1 PAV75790.1 LIAE01007639 PAV77912.1 GDIQ01102589 JAL49137.1 GDIP01067606 JAM36109.1 GDIQ01104425 JAL47301.1 FOMX01000026 SFF04605.1 LFJK02000005 POM49163.1 PVNL01000019 PRQ09509.1 PVNK01000015 PRQ05323.1 JMCC02000011 KIG18352.1 MGXV01000089 OGT34925.1 LKCN02000007 RCI12967.1 PDHQ01000162 RDA90277.1 JPKC01000171 KFZ01618.1 LUKN01002828 OAQ98532.1 AZHF01000007 OAA73049.1 PDHP01000033 RDA86842.1 AZIL01000038 EWM30282.1 KQ964783 KXN66002.1 MCFE01000445 ORX89423.1 CP023322 ATY58678.1 LSYS01005497 OPJ77037.1 MUZQ01000109 OWK58100.1 QUSF01000010 RLW05965.1 AGCU01155374 AGCU01155375 AGCU01155376 AADN05000647 AJ720832 CAG32491.1 QRBI01000117 RMC08720.1 KL671386 KFW82046.1 KB535465 EMP33601.1 MUGK01000090 OOO01917.1 LJTN01000014 KPK16474.1
PZC76211.1 KQ461184 KPJ07522.1 KQ459449 KPJ00888.1 AGBW02010741 OWR47932.1 JTDY01002142 KOB72016.1 KQ971352 EFA07314.1 GALX01003323 JAB65143.1 GDIQ01184362 JAK67363.1 GDIP01052309 JAM51406.1 GDIQ01249136 JAK02589.1 GDIQ01205159 JAK46566.1 GDIP01050711 JAM53004.1 GDIP01070601 LRGB01002994 JAM33114.1 KZS05093.1 LJIJ01000081 ODN03240.1 GQ199603 GL732567 ACT20728.1 EFX76668.1 GDIP01088803 JAM14912.1 GDIP01046912 JAM56803.1 GDIP01019387 JAM84328.1 GDIQ01133836 JAL17890.1 GECU01008590 JAS99116.1 NIVC01000721 PAA77819.1 LNIX01000001 OXA63121.1 NIVC01000017 PAA94059.1 NIVC01000720 PAA77842.1 GEDC01027191 JAS10107.1 QCYY01000619 ROT83942.1 GDIQ01149832 JAL01894.1 GDIP01171442 JAJ51960.1 GDRN01062447 JAI65069.1 DS268450 EFP03687.1 BX284606 CAA22461.1 NIPN01000002 OZG23886.1 EAAA01002802 GL379788 EGT40606.1 PDUG01000006 PIC20596.1 HE601041 CAP27409.1 GDIP01171444 JAJ51958.1 GDRN01062450 JAI65068.1 GDIQ01019818 JAN74919.1 LIAE01008038 PAV75789.1 NMWX01000001 OZG07670.1 GDIP01181780 JAJ41622.1 PAV75790.1 LIAE01007639 PAV77912.1 GDIQ01102589 JAL49137.1 GDIP01067606 JAM36109.1 GDIQ01104425 JAL47301.1 FOMX01000026 SFF04605.1 LFJK02000005 POM49163.1 PVNL01000019 PRQ09509.1 PVNK01000015 PRQ05323.1 JMCC02000011 KIG18352.1 MGXV01000089 OGT34925.1 LKCN02000007 RCI12967.1 PDHQ01000162 RDA90277.1 JPKC01000171 KFZ01618.1 LUKN01002828 OAQ98532.1 AZHF01000007 OAA73049.1 PDHP01000033 RDA86842.1 AZIL01000038 EWM30282.1 KQ964783 KXN66002.1 MCFE01000445 ORX89423.1 CP023322 ATY58678.1 LSYS01005497 OPJ77037.1 MUZQ01000109 OWK58100.1 QUSF01000010 RLW05965.1 AGCU01155374 AGCU01155375 AGCU01155376 AADN05000647 AJ720832 CAG32491.1 QRBI01000117 RMC08720.1 KL671386 KFW82046.1 KB535465 EMP33601.1 MUGK01000090 OOO01917.1 LJTN01000014 KPK16474.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000007266 UP000192223 UP000076858 UP000094527 UP000000305 UP000215902 UP000198287 UP000283509 UP000008281 UP000001940 UP000216463 UP000008144 UP000007875 UP000005237 UP000095282 UP000008068 UP000095284 UP000230233 UP000008549 UP000005239 UP000218231 UP000216624 UP000199400 UP000237256 UP000238823 UP000237968 UP000031599 UP000253664 UP000253071 UP000029284 UP000076881 UP000252748 UP000070444 UP000193498 UP000190648 UP000197619 UP000276834 UP000007267 UP000000539 UP000001646 UP000269221 UP000053258 UP000031443 UP000191121
UP000007266 UP000192223 UP000076858 UP000094527 UP000000305 UP000215902 UP000198287 UP000283509 UP000008281 UP000001940 UP000216463 UP000008144 UP000007875 UP000005237 UP000095282 UP000008068 UP000095284 UP000230233 UP000008549 UP000005239 UP000218231 UP000216624 UP000199400 UP000237256 UP000238823 UP000237968 UP000031599 UP000253664 UP000253071 UP000029284 UP000076881 UP000252748 UP000070444 UP000193498 UP000190648 UP000197619 UP000276834 UP000007267 UP000000539 UP000001646 UP000269221 UP000053258 UP000031443 UP000191121
PRIDE
Pfam
PF01565 FAD_binding_4
Interpro
Gene 3D
ProteinModelPortal
H9JSW2
A0A2A4JYM3
A0A2H1VSB6
A0A2W1BPJ6
A0A0N1ID81
A0A0N1PGV7
+ More
A0A212F2H7 A0A0L7L9T6 D6WTX2 V5GXQ9 A0A1W4WBU7 A0A0P5KUL6 A0A0P5YW80 A0A0P5GEV7 A0A0P5J9A1 A0A0N8DB92 A0A0P5YK95 A0A1D2NDB4 C7DR07 A0A0P5WAM6 A0A0P6A5I7 A0A0P6BAP2 A0A0P5PHY4 A0A1B6JIJ3 A0A267FXT9 A0A226F1P3 A0A267H929 A0A267FY07 A0A1B6C9D4 A0A3R7T0B8 A0A0P5NP72 A0A0P5CP71 A0A1W2W6U8 A0A0P4WH00 E3MJD7 Q9XVZ2 A0A261CPQ7 F6ZXA7 H2ZIN9 A0A2H2I517 A0A1I7TRT2 G0MAQ7 A0A1I7SVI7 A0A2G5T0E7 A8X449 A0A0P5CH18 A0A0P4W8Z8 A0A0P6HFF3 H3EYA8 A0A2A2KPH2 A0A261BD58 A0A0P5BQE7 A0A2A2KPD6 A0A2A2KVH6 A0A0P5RHF8 A0A0P5XTY9 A0A0P5RPZ0 A0A1I2FH59 A0A2P4WHZ1 A0A2S9YWS8 A0A2S9YJN9 A0A0C2DFF7 A0A1G0GBE5 A0A367LEX4 A0A369H047 A0A094IY66 A0A179I8B4 A0A162MZF9 A0A369GVF9 W7TVX8 A0A137NTE9 A0A1Y1XUM5 A0A2H4S6G3 A0A1V4JZG3 A0A218UXF9 A0A3L8SQF0 K7F6B9 Q5ZIF2 G1K8B5 A0A3M0KCB9 A0A093Q065 M7B6K0 A0A1S9CYJ4 A0A0S8BZW2
A0A212F2H7 A0A0L7L9T6 D6WTX2 V5GXQ9 A0A1W4WBU7 A0A0P5KUL6 A0A0P5YW80 A0A0P5GEV7 A0A0P5J9A1 A0A0N8DB92 A0A0P5YK95 A0A1D2NDB4 C7DR07 A0A0P5WAM6 A0A0P6A5I7 A0A0P6BAP2 A0A0P5PHY4 A0A1B6JIJ3 A0A267FXT9 A0A226F1P3 A0A267H929 A0A267FY07 A0A1B6C9D4 A0A3R7T0B8 A0A0P5NP72 A0A0P5CP71 A0A1W2W6U8 A0A0P4WH00 E3MJD7 Q9XVZ2 A0A261CPQ7 F6ZXA7 H2ZIN9 A0A2H2I517 A0A1I7TRT2 G0MAQ7 A0A1I7SVI7 A0A2G5T0E7 A8X449 A0A0P5CH18 A0A0P4W8Z8 A0A0P6HFF3 H3EYA8 A0A2A2KPH2 A0A261BD58 A0A0P5BQE7 A0A2A2KPD6 A0A2A2KVH6 A0A0P5RHF8 A0A0P5XTY9 A0A0P5RPZ0 A0A1I2FH59 A0A2P4WHZ1 A0A2S9YWS8 A0A2S9YJN9 A0A0C2DFF7 A0A1G0GBE5 A0A367LEX4 A0A369H047 A0A094IY66 A0A179I8B4 A0A162MZF9 A0A369GVF9 W7TVX8 A0A137NTE9 A0A1Y1XUM5 A0A2H4S6G3 A0A1V4JZG3 A0A218UXF9 A0A3L8SQF0 K7F6B9 Q5ZIF2 G1K8B5 A0A3M0KCB9 A0A093Q065 M7B6K0 A0A1S9CYJ4 A0A0S8BZW2
PDB
5L6G
E-value=0.000197982,
Score=108
Ontologies
PATHWAY
GO
GO:0016491
GO:0071949
GO:0016021
GO:0050660
GO:0008202
GO:0016628
GO:0016020
GO:0005737
GO:0000246
GO:0019899
GO:0031639
GO:0030539
GO:0009888
GO:0006979
GO:0005783
GO:0043588
GO:0043154
GO:0005634
GO:0042987
GO:0008104
GO:0061024
GO:0042605
GO:0006695
GO:0003824
GO:0016614
GO:0006508
GO:0005515
GO:0006811
GO:0005622
GO:0006810
GO:0055114
PANTHER
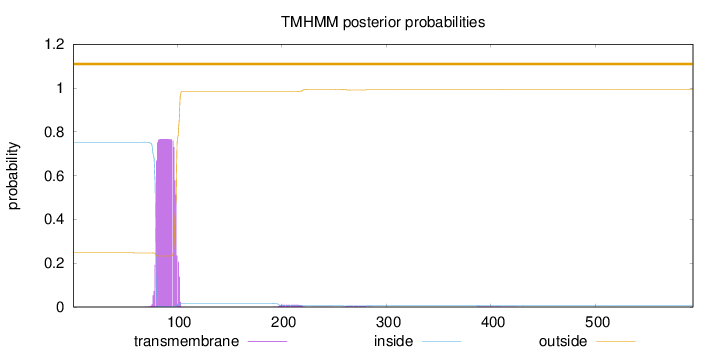
Topology
Length:
593
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
16.06728
Exp number, first 60 AAs:
0.00026
Total prob of N-in:
0.75277
outside
1 - 593
Population Genetic Test Statistics
Pi
150.289218
Theta
208.252144
Tajima's D
-0.88062
CLR
1.10772
CSRT
0.163541822908855
Interpretation
Uncertain
