Gene
KWMTBOMO11250
Pre Gene Modal
BGIBMGA012623
Annotation
PREDICTED:_general_transcription_factor_IIH_subunit_4-like_[Plutella_xylostella]
Full name
General transcription factor IIH subunit 4
Location in the cell
Mitochondrial Reliability : 1.6 PlasmaMembrane Reliability : 1.252
Sequence
CDS
ATGTCAGAGTCTTCTAAATCGAAGTCTTTAAATTTGAATCCTTCGCCTACACTACAATGCAAAGACCTTCACGAATATCTTAAGTCGCGGTCTCCGCAGTTTCTAGAGACTCTTTACAATTACCCCACTATATGTCTCGCTGTATATCGCGAATTACCAGAGCTAGCACGTCACTTCGTAATCCGCTTGCTGTTCGTTGAGCAGCCAGTCCCACAGGCAGTTGTCACATCCTGGGTCTCGCAAACATATGCCAAAGAACAAGCAAAAGCATGCACTACACTGTCAGAGCTCTCCGTGTGGCAGGAGGCCCCCATACCGGGGGGCATGCCGGGCTGGATGCTCGCACAGTCGTTTAAAAAGAACCTCAAAGTGTCATTGCTTGGGGGAGGCAGGGCGTGGAGCATGTCGTCGTCGCTGGAGCCGGACGGGAAGGCCCGGGACGTGTCGTTCCTGGACGCGTACGCGCTGGAGCGCTGGGAGTGCGTGCTGCACTACATGGTGGGCAGCGCGCAGACCGAGGGCATCAGCGCCGACGCCGTCCGCATACTGCTGCACGCCGGACTCATGACCAGAGACGCAGAAGACGGGTCCGCCGTTATCACTCGCGCCGGCTTCCAGTTTCTGCTCCTGAGTACACCCAAGCAGGTGTGGCTGTTCCTGCAGCACTACCTGCACACGGCCGAGCGCCGCAGCCTCAGCGCCGCCGAGTGCCTGGCCTTCCTCTACCAGCTCAGCTTCAGCACCCTGGGGAAGGACTACAGCACGGAGGGCATGAGCAACAACATGCTGGTGTTCCTGCAGCACCTGCGGGAGTTCGGCCTCGTCTACCAGAGGAAGCGCAAGGCGGGCCGCTTCTACCCCACCCGCCTCGCGCTCAACATCCTGCGCGTGCGCTGCGCCGCGCCGCTCGAGGCCGCCGCCGGCGCGCGCGGACACCTCGTGCTGGAGACCAACTACCGCGTGTACGGCTACACCTCCTCCGACCTGCAGGTCGCGCTGCTCGGCCTCTTCACGGAGCTCATCTACAGGTTCCCCAACGTGGTGGTGGGCGTGGTGAGCCGGGAGTCCGTCCGCCAGGCGCTGCGCGGTGGCATCACGGCCGAGCAGATCATCCACTACCTGGAGCAGCACGCGCACCCGCAGATGCTGAAGTCAGAGACGGGCGGCATCCGGTCGGCGTCGATGCTGCCCCCCACCGTGGTGGACCAGATCCGCCTGTGGGAGTCCGAGAGGAACCGCTGCGTCTACACGGAGGGGGTCGTCTACAACCAGTTCCTGTCGCAGGCCGACTTCGTGGTGCTGCGGGACCACGCCCGCGCGGCCGGGGTGCTGAGCTGGCAGCACGAGCGCGCGCGGACCATGGTGGTGACCCGCGCCGGACACGACGACGTCAAGAAGTTCTGGAAGCGCTACTCCAAGAACAGCTAG
Protein
MSESSKSKSLNLNPSPTLQCKDLHEYLKSRSPQFLETLYNYPTICLAVYRELPELARHFVIRLLFVEQPVPQAVVTSWVSQTYAKEQAKACTTLSELSVWQEAPIPGGMPGWMLAQSFKKNLKVSLLGGGRAWSMSSSLEPDGKARDVSFLDAYALERWECVLHYMVGSAQTEGISADAVRILLHAGLMTRDAEDGSAVITRAGFQFLLLSTPKQVWLFLQHYLHTAERRSLSAAECLAFLYQLSFSTLGKDYSTEGMSNNMLVFLQHLREFGLVYQRKRKAGRFYPTRLALNILRVRCAAPLEAAAGARGHLVLETNYRVYGYTSSDLQVALLGLFTELIYRFPNVVVGVVSRESVRQALRGGITAEQIIHYLEQHAHPQMLKSETGGIRSASMLPPTVVDQIRLWESERNRCVYTEGVVYNQFLSQADFVVLRDHARAAGVLSWQHERARTMVVTRAGHDDVKKFWKRYSKNS
Summary
Description
Component of the general transcription and DNA repair factor IIH (TFIIH) core complex which is involved in general and transcription-coupled nucleotide excision repair (NER) of damaged DNA.
Similarity
Belongs to the TFB2 family.
Uniprot
H9JSW1
A0A2W1BMC3
A0A212F2G2
A0A2A4JXQ6
A0A2H1VRK8
A0A1W4XA12
+ More
A0A1Y1LPE6 A0A0N1ICW4 A0A182RKF3 A0A182UFC4 D6WU09 A0A182WUK1 A0A182FTP8 A0A182VHI5 A0A182KKF9 Q7QH12 A0A182HVX2 A0A182NZX6 A0A182MCE1 A0A182N2Z6 W5JH20 A0A182IRW5 A0A182JV50 A0A182W221 A0A182YBM3 A0A2M4AHE0 A0A182Q025 A0A195ER83 A0A067R066 A0A182HD83 E2B8N7 Q176J9 A0A154PDQ4 A0A0M8ZZ27 E2AJG8 A0A1B0GJ18 A0A151WNZ1 A0A195DMQ2 A0A195AVR0 A0A151I7U4 A0A087ZX20 A0A158P1W3 A0A026WC09 A0A2A3EBW6 A0A1Q3F3U2 B0WFU1 A0A084W999 W8C5B7 K7JBE3 A0A1I8MA08 A0A1I8P8E8 A0A2P8ZEZ1 A0A0L0CDV9 A0A240SWY5 A0A1B0AR15 A0A1A9Z8Z4 A0A1A9YG33 A0A034VQF4 A0A1A9V4D4 A0A1B6MGU7 U4UR34 E0VYT4 N6TTI5 A0A336K8B7 A0A1B6H6W5 A0A1B6C308 A0A310SPQ2 B3MAD1 B4KUK0 B4LH90 A0A1W4VNC4 A0A3B0KWK2 B4N4V6 Q2M1D2 B4GUA2 B4IWU6 A0A0A9Y7B7 B3NI43 B4ITX7 A0A0J9RVN7 B4HIG7 Q9VUR1 A0A0P4VQN4 F4W639 A0A069DU73 A0A0V0G7D6 A0A224XC95 B4QL39 T1HG45 A0A182TUF5 A0A0L7R9A6 A0A023F391 A0A0M4EBB5 A0A2R7WBZ7 L7M5B7 A0A224ZAR2 A0A131YRK0 A0A1E1XG20 A0A131Y1W6 A0A2R5LGB0
A0A1Y1LPE6 A0A0N1ICW4 A0A182RKF3 A0A182UFC4 D6WU09 A0A182WUK1 A0A182FTP8 A0A182VHI5 A0A182KKF9 Q7QH12 A0A182HVX2 A0A182NZX6 A0A182MCE1 A0A182N2Z6 W5JH20 A0A182IRW5 A0A182JV50 A0A182W221 A0A182YBM3 A0A2M4AHE0 A0A182Q025 A0A195ER83 A0A067R066 A0A182HD83 E2B8N7 Q176J9 A0A154PDQ4 A0A0M8ZZ27 E2AJG8 A0A1B0GJ18 A0A151WNZ1 A0A195DMQ2 A0A195AVR0 A0A151I7U4 A0A087ZX20 A0A158P1W3 A0A026WC09 A0A2A3EBW6 A0A1Q3F3U2 B0WFU1 A0A084W999 W8C5B7 K7JBE3 A0A1I8MA08 A0A1I8P8E8 A0A2P8ZEZ1 A0A0L0CDV9 A0A240SWY5 A0A1B0AR15 A0A1A9Z8Z4 A0A1A9YG33 A0A034VQF4 A0A1A9V4D4 A0A1B6MGU7 U4UR34 E0VYT4 N6TTI5 A0A336K8B7 A0A1B6H6W5 A0A1B6C308 A0A310SPQ2 B3MAD1 B4KUK0 B4LH90 A0A1W4VNC4 A0A3B0KWK2 B4N4V6 Q2M1D2 B4GUA2 B4IWU6 A0A0A9Y7B7 B3NI43 B4ITX7 A0A0J9RVN7 B4HIG7 Q9VUR1 A0A0P4VQN4 F4W639 A0A069DU73 A0A0V0G7D6 A0A224XC95 B4QL39 T1HG45 A0A182TUF5 A0A0L7R9A6 A0A023F391 A0A0M4EBB5 A0A2R7WBZ7 L7M5B7 A0A224ZAR2 A0A131YRK0 A0A1E1XG20 A0A131Y1W6 A0A2R5LGB0
Pubmed
19121390
28756777
22118469
28004739
26354079
18362917
+ More
19820115 20966253 12364791 14747013 17210077 20920257 23761445 25244985 24845553 26483478 20798317 17510324 21347285 24508170 30249741 24438588 24495485 20075255 25315136 29403074 26108605 25348373 23537049 20566863 17994087 15632085 25401762 26823975 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 27129103 21719571 26334808 25474469 25576852 28797301 26830274 28503490
19820115 20966253 12364791 14747013 17210077 20920257 23761445 25244985 24845553 26483478 20798317 17510324 21347285 24508170 30249741 24438588 24495485 20075255 25315136 29403074 26108605 25348373 23537049 20566863 17994087 15632085 25401762 26823975 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 27129103 21719571 26334808 25474469 25576852 28797301 26830274 28503490
EMBL
BABH01035991
BABH01035992
BABH01035993
KZ149965
PZC76209.1
AGBW02010741
+ More
OWR47930.1 NWSH01000411 PCG76589.1 ODYU01003846 SOQ43112.1 GEZM01050589 GEZM01050588 JAV75509.1 KQ459449 KPJ00890.1 KQ971352 EFA07302.1 AAAB01008823 EAA05440.1 APCN01001435 AXCM01000173 ADMH02001207 ETN63662.1 GGFK01006882 MBW40203.1 AXCN02001144 KQ982021 KYN30417.1 KK852804 KDR16235.1 JXUM01129107 KQ567403 KXJ69446.1 GL446361 EFN87943.1 CH477386 EAT42083.1 KQ434879 KZC09983.1 KQ435794 KOX74030.1 GL439992 EFN66420.1 AJWK01019210 KQ982892 KYQ49622.1 KQ980724 KYN14153.1 KQ976731 KYM76328.1 KQ978397 KYM94206.1 ADTU01006900 KK107293 QOIP01000005 EZA53181.1 RLU22624.1 KZ288309 PBC28676.1 GFDL01012856 JAV22189.1 DS231920 EDS26435.1 ATLV01021659 KE525321 KFB46793.1 GAMC01009121 JAB97434.1 AAZX01015659 PYGN01000077 PSN55058.1 JRES01000502 KNC30598.1 CCAG010006155 JXJN01002206 GAKP01014902 JAC44050.1 GEBQ01004845 JAT35132.1 KB632331 ERL92616.1 DS235847 EEB18540.1 APGK01019264 KB740098 ENN81378.1 UFQS01000174 UFQT01000174 SSX00731.1 SSX21111.1 GECU01037270 JAS70436.1 GEDC01029390 JAS07908.1 KQ761323 OAD57894.1 CH902618 EDV40182.1 CH933809 EDW18228.2 CH940647 EDW70603.1 OUUW01000027 SPP89741.1 CH964101 EDW79395.1 CH379069 EAL30642.3 CH479191 EDW26185.1 CH916366 EDV97347.1 GBHO01016621 GBRD01002397 GDHC01016762 JAG26983.1 JAG63424.1 JAQ01867.1 CH954178 EDV52129.1 CH891737 EDW99840.1 CM002912 KMY99778.1 CH480815 EDW41597.1 AE014296 AAF49614.1 GDKW01001805 JAI54790.1 GL887707 EGI70174.1 GBGD01001414 JAC87475.1 GECL01002239 JAP03885.1 GFTR01006391 JAW10035.1 CM000363 EDX10564.1 ACPB03001139 KQ414628 KOC67341.1 GBBI01002989 JAC15723.1 CP012525 ALC44716.1 KK854594 PTY17214.1 GACK01006745 JAA58289.1 GFPF01012636 MAA23782.1 GEDV01006674 JAP81883.1 GFAC01000996 JAT98192.1 GEFM01002919 JAP72877.1 GGLE01004437 MBY08563.1
OWR47930.1 NWSH01000411 PCG76589.1 ODYU01003846 SOQ43112.1 GEZM01050589 GEZM01050588 JAV75509.1 KQ459449 KPJ00890.1 KQ971352 EFA07302.1 AAAB01008823 EAA05440.1 APCN01001435 AXCM01000173 ADMH02001207 ETN63662.1 GGFK01006882 MBW40203.1 AXCN02001144 KQ982021 KYN30417.1 KK852804 KDR16235.1 JXUM01129107 KQ567403 KXJ69446.1 GL446361 EFN87943.1 CH477386 EAT42083.1 KQ434879 KZC09983.1 KQ435794 KOX74030.1 GL439992 EFN66420.1 AJWK01019210 KQ982892 KYQ49622.1 KQ980724 KYN14153.1 KQ976731 KYM76328.1 KQ978397 KYM94206.1 ADTU01006900 KK107293 QOIP01000005 EZA53181.1 RLU22624.1 KZ288309 PBC28676.1 GFDL01012856 JAV22189.1 DS231920 EDS26435.1 ATLV01021659 KE525321 KFB46793.1 GAMC01009121 JAB97434.1 AAZX01015659 PYGN01000077 PSN55058.1 JRES01000502 KNC30598.1 CCAG010006155 JXJN01002206 GAKP01014902 JAC44050.1 GEBQ01004845 JAT35132.1 KB632331 ERL92616.1 DS235847 EEB18540.1 APGK01019264 KB740098 ENN81378.1 UFQS01000174 UFQT01000174 SSX00731.1 SSX21111.1 GECU01037270 JAS70436.1 GEDC01029390 JAS07908.1 KQ761323 OAD57894.1 CH902618 EDV40182.1 CH933809 EDW18228.2 CH940647 EDW70603.1 OUUW01000027 SPP89741.1 CH964101 EDW79395.1 CH379069 EAL30642.3 CH479191 EDW26185.1 CH916366 EDV97347.1 GBHO01016621 GBRD01002397 GDHC01016762 JAG26983.1 JAG63424.1 JAQ01867.1 CH954178 EDV52129.1 CH891737 EDW99840.1 CM002912 KMY99778.1 CH480815 EDW41597.1 AE014296 AAF49614.1 GDKW01001805 JAI54790.1 GL887707 EGI70174.1 GBGD01001414 JAC87475.1 GECL01002239 JAP03885.1 GFTR01006391 JAW10035.1 CM000363 EDX10564.1 ACPB03001139 KQ414628 KOC67341.1 GBBI01002989 JAC15723.1 CP012525 ALC44716.1 KK854594 PTY17214.1 GACK01006745 JAA58289.1 GFPF01012636 MAA23782.1 GEDV01006674 JAP81883.1 GFAC01000996 JAT98192.1 GEFM01002919 JAP72877.1 GGLE01004437 MBY08563.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000192223
UP000053268
UP000075900
+ More
UP000075902 UP000007266 UP000076407 UP000069272 UP000075903 UP000075882 UP000007062 UP000075840 UP000075885 UP000075883 UP000075884 UP000000673 UP000075880 UP000075881 UP000075920 UP000076408 UP000075886 UP000078541 UP000027135 UP000069940 UP000249989 UP000008237 UP000008820 UP000076502 UP000053105 UP000000311 UP000092461 UP000075809 UP000078492 UP000078540 UP000078542 UP000005203 UP000005205 UP000053097 UP000279307 UP000242457 UP000002320 UP000030765 UP000002358 UP000095301 UP000095300 UP000245037 UP000037069 UP000092444 UP000092460 UP000092445 UP000092443 UP000078200 UP000030742 UP000009046 UP000019118 UP000007801 UP000009192 UP000008792 UP000192221 UP000268350 UP000007798 UP000001819 UP000008744 UP000001070 UP000008711 UP000002282 UP000001292 UP000000803 UP000007755 UP000000304 UP000015103 UP000053825 UP000092553
UP000075902 UP000007266 UP000076407 UP000069272 UP000075903 UP000075882 UP000007062 UP000075840 UP000075885 UP000075883 UP000075884 UP000000673 UP000075880 UP000075881 UP000075920 UP000076408 UP000075886 UP000078541 UP000027135 UP000069940 UP000249989 UP000008237 UP000008820 UP000076502 UP000053105 UP000000311 UP000092461 UP000075809 UP000078492 UP000078540 UP000078542 UP000005203 UP000005205 UP000053097 UP000279307 UP000242457 UP000002320 UP000030765 UP000002358 UP000095301 UP000095300 UP000245037 UP000037069 UP000092444 UP000092460 UP000092445 UP000092443 UP000078200 UP000030742 UP000009046 UP000019118 UP000007801 UP000009192 UP000008792 UP000192221 UP000268350 UP000007798 UP000001819 UP000008744 UP000001070 UP000008711 UP000002282 UP000001292 UP000000803 UP000007755 UP000000304 UP000015103 UP000053825 UP000092553
ProteinModelPortal
H9JSW1
A0A2W1BMC3
A0A212F2G2
A0A2A4JXQ6
A0A2H1VRK8
A0A1W4XA12
+ More
A0A1Y1LPE6 A0A0N1ICW4 A0A182RKF3 A0A182UFC4 D6WU09 A0A182WUK1 A0A182FTP8 A0A182VHI5 A0A182KKF9 Q7QH12 A0A182HVX2 A0A182NZX6 A0A182MCE1 A0A182N2Z6 W5JH20 A0A182IRW5 A0A182JV50 A0A182W221 A0A182YBM3 A0A2M4AHE0 A0A182Q025 A0A195ER83 A0A067R066 A0A182HD83 E2B8N7 Q176J9 A0A154PDQ4 A0A0M8ZZ27 E2AJG8 A0A1B0GJ18 A0A151WNZ1 A0A195DMQ2 A0A195AVR0 A0A151I7U4 A0A087ZX20 A0A158P1W3 A0A026WC09 A0A2A3EBW6 A0A1Q3F3U2 B0WFU1 A0A084W999 W8C5B7 K7JBE3 A0A1I8MA08 A0A1I8P8E8 A0A2P8ZEZ1 A0A0L0CDV9 A0A240SWY5 A0A1B0AR15 A0A1A9Z8Z4 A0A1A9YG33 A0A034VQF4 A0A1A9V4D4 A0A1B6MGU7 U4UR34 E0VYT4 N6TTI5 A0A336K8B7 A0A1B6H6W5 A0A1B6C308 A0A310SPQ2 B3MAD1 B4KUK0 B4LH90 A0A1W4VNC4 A0A3B0KWK2 B4N4V6 Q2M1D2 B4GUA2 B4IWU6 A0A0A9Y7B7 B3NI43 B4ITX7 A0A0J9RVN7 B4HIG7 Q9VUR1 A0A0P4VQN4 F4W639 A0A069DU73 A0A0V0G7D6 A0A224XC95 B4QL39 T1HG45 A0A182TUF5 A0A0L7R9A6 A0A023F391 A0A0M4EBB5 A0A2R7WBZ7 L7M5B7 A0A224ZAR2 A0A131YRK0 A0A1E1XG20 A0A131Y1W6 A0A2R5LGB0
A0A1Y1LPE6 A0A0N1ICW4 A0A182RKF3 A0A182UFC4 D6WU09 A0A182WUK1 A0A182FTP8 A0A182VHI5 A0A182KKF9 Q7QH12 A0A182HVX2 A0A182NZX6 A0A182MCE1 A0A182N2Z6 W5JH20 A0A182IRW5 A0A182JV50 A0A182W221 A0A182YBM3 A0A2M4AHE0 A0A182Q025 A0A195ER83 A0A067R066 A0A182HD83 E2B8N7 Q176J9 A0A154PDQ4 A0A0M8ZZ27 E2AJG8 A0A1B0GJ18 A0A151WNZ1 A0A195DMQ2 A0A195AVR0 A0A151I7U4 A0A087ZX20 A0A158P1W3 A0A026WC09 A0A2A3EBW6 A0A1Q3F3U2 B0WFU1 A0A084W999 W8C5B7 K7JBE3 A0A1I8MA08 A0A1I8P8E8 A0A2P8ZEZ1 A0A0L0CDV9 A0A240SWY5 A0A1B0AR15 A0A1A9Z8Z4 A0A1A9YG33 A0A034VQF4 A0A1A9V4D4 A0A1B6MGU7 U4UR34 E0VYT4 N6TTI5 A0A336K8B7 A0A1B6H6W5 A0A1B6C308 A0A310SPQ2 B3MAD1 B4KUK0 B4LH90 A0A1W4VNC4 A0A3B0KWK2 B4N4V6 Q2M1D2 B4GUA2 B4IWU6 A0A0A9Y7B7 B3NI43 B4ITX7 A0A0J9RVN7 B4HIG7 Q9VUR1 A0A0P4VQN4 F4W639 A0A069DU73 A0A0V0G7D6 A0A224XC95 B4QL39 T1HG45 A0A182TUF5 A0A0L7R9A6 A0A023F391 A0A0M4EBB5 A0A2R7WBZ7 L7M5B7 A0A224ZAR2 A0A131YRK0 A0A1E1XG20 A0A131Y1W6 A0A2R5LGB0
PDB
6O9M
E-value=5.03979e-134,
Score=1225
Ontologies
PATHWAY
GO
PANTHER
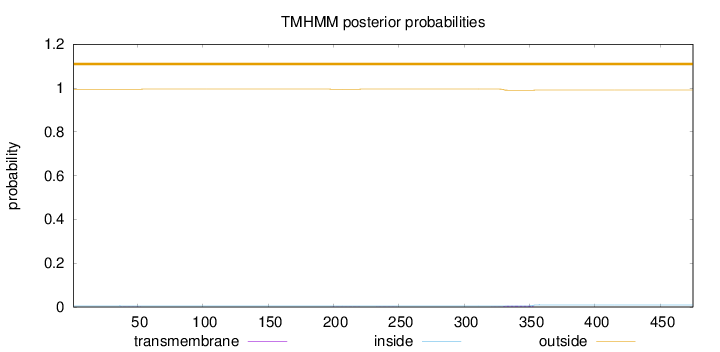
Topology
Subcellular location
Nucleus
Length:
475
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.13642
Exp number, first 60 AAs:
0.00321
Total prob of N-in:
0.00500
outside
1 - 475
Population Genetic Test Statistics
Pi
173.363522
Theta
146.168144
Tajima's D
0.399532
CLR
0.076116
CSRT
0.489125543722814
Interpretation
Uncertain
