Gene
KWMTBOMO11247
Pre Gene Modal
BGIBMGA012620
Annotation
PREDICTED:_protein_prickle-like_[Bombyx_mori]
Full name
Metalloendopeptidase
+ More
Protein prickle
Protein espinas
Protein prickle
Protein espinas
Alternative Name
Protein spiny legs
Location in the cell
Nuclear Reliability : 2.617
Sequence
CDS
ATGTCGCTATCCCCCCATCCAGTGCCTCAAGGGAAGTTCAGGGGTCTACTAGAACACGCTGAACGGTTGATGAAACCAGTGGATCCTCATCGGCATTCTCAAAGCGACGACGATTCCGGATGTGCTTTGGAAGAGTATACGTGGGTTCCGCCTGGATTGAGACCAGAACAGGTCCATCTTTACTTCTCAGCACTTCCGGAAGACAAGGTCCCGTACGTGAATAGCGTAGGGGAGAGGTACCGCCTCAAGCAACTACTTCAACAACTTCCACCTCAAGATAATGAGGTTCGGTACTGCCATGCTTTATCTGATGAAGAGAGGAAGGAGTTGAGGCTGTTCAGCGCACAGAGGAAACGCGAGGCCCTGGGTAGAGGACAGGCGAGGCAGCTCCACGCATCGGCGCCTTGCGAGAGGAGTGGATCGTGA
Protein
MSLSPHPVPQGKFRGLLEHAERLMKPVDPHRHSQSDDDSGCALEEYTWVPPGLRPEQVHLYFSALPEDKVPYVNSVGERYRLKQLLQQLPPQDNEVRYCHALSDEERKELRLFSAQRKREALGRGQARQLHASAPCERSGS
Summary
Description
Acts in a planar cell polarity (PCP) complex; polarization along the apical/basal axis of epithelial cells. PCP signaling in the wing disk requires the receptor fz and the cytoplasmic proteins dsh and pk. These act in a feedback loop leading to activation of the jnk cascade and subsequent polarized arrangement of hairs and bristles. Dgo and pk compete with one another for dsh binding, thereby modulating fz dsh activity and ensuring tight control over fz PCP signaling. Vang, stan and pk function together to regulate the establishment of tissue polarity in the adult eye (By similarity).
Acts in a planar cell polarity (PCP) complex; polarization along the apical/basal axis of epithelial cells. Correct expression of the alternative isoforms is required for PCP signaling in imaginal disks. PCP signaling in the wing disk requires the receptor fz and the cytoplasmic proteins dsh and pk. These act in a feedback loop leading to activation of the jnk cascade and subsequent polarized arrangement of hairs and bristles. Dgo and pk compete with one another for dsh binding, thereby modulating fz dsh activity and ensuring tight control over fz PCP signaling. Vang, stan and pk function together to regulate the establishment of tissue polarity in the adult eye.
Acts in a planar cell polarity (PCP) complex; polarization along the apical/basal axis of epithelial cells. Correct expression of the alternative isoforms is required for PCP signaling in imaginal disks. PCP signaling in the wing disk requires the receptor fz and the cytoplasmic proteins dsh and pk. These act in a feedback loop leading to activation of the jnk cascade and subsequent polarized arrangement of hairs and bristles. Dgo and pk compete with one another for dsh binding, thereby modulating fz dsh activity and ensuring tight control over fz PCP signaling. Vang, stan and pk function together to regulate the establishment of tissue polarity in the adult eye.
Cofactor
Zn(2+)
Subunit
Interacts with dsh; PET and LIM domains interact with dsh DEP domain, in wing cells. Interacts with Vang in photoreceptor cells (By similarity).
Interacts with dsh; PET and LIM domains interact with dsh DEP domain, in wing cells. Interacts with Vang in photoreceptor cells.
Interacts with dsh; PET and LIM domains interact with dsh DEP domain, in wing cells. Interacts with Vang in photoreceptor cells.
Similarity
Belongs to the prickle / espinas / testin family.
Keywords
Cell membrane
Complete proteome
Developmental protein
LIM domain
Membrane
Metal-binding
Reference proteome
Repeat
Zinc
Alternative splicing
Feature
chain Protein prickle
splice variant In isoform B.
splice variant In isoform B.
Uniprot
A0A2H1V5P8
A0A212F2G1
A0A3S2NPD8
A0A2A4JX68
A0A2W1BSM4
H9JSV8
+ More
A0A0N0P9S6 A0A1B6MDH1 A0A1B6GXL9 A0A1W4WID4 A0A1B6DJX5 D6WPW4 U4U4V9 A0A336K203 A0A336LMA4 A0A336K8B4 A0A1Y1NDG2 W5JT26 A0A182FIB0 A0A2M4AG89 A0A2M4AFY9 A0A182TBX7 A0A1B0AYM5 A0A2M4AEA4 A0A2M4AEI1 A0A2M4AE79 A0A182WIT6 A0A182QDT9 A0A2S2NDT6 A0A182MN04 A0A182NHP1 A0A182P9V8 A0A182RFQ4 A0A1A9W160 A0A034V1S2 B4QE78 A0A182XZH5 A0A1B0AAU4 A0A1J1I8E1 A0A182X6G2 Q7QJT4 A0A182I3R6 A0A182JEV1 A0A182KRW2 A0A0L7LM11 A0A2P2HYN0 A0A2R7X3E7 E9G6K1 A0A0A1X7J1 B4GCL9 A0A2S2N8J4 A0A2S2QRT3 B4J3Z0 A0A0K8TYQ3 B4MPX6 A0A1I8MQV6 Q292U5 A1Z6W3-2 A0A2H8TJM4 F3YDG0 A0A0Q5WPJ4 B4QDW0 E2QC70 Q9U1I1 B3NAF9 A1Z6W3 A1Z6W3-3 J9JLZ2 A0A3B0JI68 B4HQQ2 B4P1E1 A0A0P6G1G6 A0A0B4KED7 B3MHQ1 A0A0B4KEK5 B4J405 A0A0J9R8C4 A0A0P5C5Q7 A0A1W4V1C8 B3NAH4 A0A0P5CI28 A0A0J9TV34 B4MFG1 A0A0P6F0X0 A0A0P5LCT1 W8BTE7 A0A0J9R8C8 A0A0P6FNR5 A0A0P6G6B4 A0A0P5G7V5 A0A1W4V210 B4GCM4 A0A0P5G751 A0A3B0IYY3 A0A0A1XE64 A0A1W4VF77 A0A0P5LHW2 A0A0R3NRM5 A0A0K8TX40
A0A0N0P9S6 A0A1B6MDH1 A0A1B6GXL9 A0A1W4WID4 A0A1B6DJX5 D6WPW4 U4U4V9 A0A336K203 A0A336LMA4 A0A336K8B4 A0A1Y1NDG2 W5JT26 A0A182FIB0 A0A2M4AG89 A0A2M4AFY9 A0A182TBX7 A0A1B0AYM5 A0A2M4AEA4 A0A2M4AEI1 A0A2M4AE79 A0A182WIT6 A0A182QDT9 A0A2S2NDT6 A0A182MN04 A0A182NHP1 A0A182P9V8 A0A182RFQ4 A0A1A9W160 A0A034V1S2 B4QE78 A0A182XZH5 A0A1B0AAU4 A0A1J1I8E1 A0A182X6G2 Q7QJT4 A0A182I3R6 A0A182JEV1 A0A182KRW2 A0A0L7LM11 A0A2P2HYN0 A0A2R7X3E7 E9G6K1 A0A0A1X7J1 B4GCL9 A0A2S2N8J4 A0A2S2QRT3 B4J3Z0 A0A0K8TYQ3 B4MPX6 A0A1I8MQV6 Q292U5 A1Z6W3-2 A0A2H8TJM4 F3YDG0 A0A0Q5WPJ4 B4QDW0 E2QC70 Q9U1I1 B3NAF9 A1Z6W3 A1Z6W3-3 J9JLZ2 A0A3B0JI68 B4HQQ2 B4P1E1 A0A0P6G1G6 A0A0B4KED7 B3MHQ1 A0A0B4KEK5 B4J405 A0A0J9R8C4 A0A0P5C5Q7 A0A1W4V1C8 B3NAH4 A0A0P5CI28 A0A0J9TV34 B4MFG1 A0A0P6F0X0 A0A0P5LCT1 W8BTE7 A0A0J9R8C8 A0A0P6FNR5 A0A0P6G6B4 A0A0P5G7V5 A0A1W4V210 B4GCM4 A0A0P5G751 A0A3B0IYY3 A0A0A1XE64 A0A1W4VF77 A0A0P5LHW2 A0A0R3NRM5 A0A0K8TX40
EC Number
3.4.24.-
Pubmed
22118469
28756777
19121390
26354079
18362917
19820115
+ More
23537049 28004739 20920257 23761445 25348373 17994087 25244985 12364791 20966253 26227816 21292972 25830018 25315136 15632085 10485852 10731132 12537572 12015986 12941693 12642492 15937478 12537568 12537573 12537574 16110336 17569856 17569867 12537569 17550304 22936249 24495485
23537049 28004739 20920257 23761445 25348373 17994087 25244985 12364791 20966253 26227816 21292972 25830018 25315136 15632085 10485852 10731132 12537572 12015986 12941693 12642492 15937478 12537568 12537573 12537574 16110336 17569856 17569867 12537569 17550304 22936249 24495485
EMBL
ODYU01000784
SOQ36096.1
AGBW02010753
OWR47914.1
RSAL01000017
RVE52914.1
+ More
NWSH01000411 PCG76591.1 KZ149965 PZC76207.1 BABH01035978 BABH01035979 BABH01035980 KQ459449 KPJ00894.1 GEBQ01005994 JAT33983.1 GECZ01002612 JAS67157.1 GEDC01011388 JAS25910.1 KQ971354 EFA06885.1 KB631648 ERL85005.1 UFQS01000067 UFQT01000067 SSW98994.1 SSX19376.1 SSW98992.1 SSX19374.1 SSW98993.1 SSX19375.1 GEZM01005749 GEZM01005748 JAV95942.1 ADMH02000160 ETN67532.1 GGFK01006475 MBW39796.1 GGFK01006379 MBW39700.1 JXJN01005875 JXJN01005876 JXJN01005877 JXJN01005878 JXJN01005879 GGFK01005759 MBW39080.1 GGFK01005868 MBW39189.1 GGFK01005772 MBW39093.1 AXCN02000591 GGMR01002668 MBY15287.1 AXCM01000153 GAKP01023256 JAC35700.1 CM000362 EDX05987.1 CVRI01000043 CRK96002.1 AAAB01008807 APCN01000218 JTDY01000684 KOB76236.1 IACF01001157 LAB66873.1 KK856713 PTY26312.1 GL732533 EFX84948.1 GBXI01007033 JAD07259.1 CH479181 EDW31467.1 GGMR01000922 MBY13541.1 GGMS01011264 MBY80467.1 CH916367 EDW01573.1 GDHF01032921 JAI19393.1 CH963849 EDW74165.1 CM000071 EAL24766.1 AJ243708 AJ243710 AJ243709 AE013599 AAF59281.2 CAB57345.3 GFXV01002542 MBW14347.1 BT126321 AEB66928.1 CH954177 KQS70970.1 EDX05972.1 AAM68907.1 AJ251892 AY094898 BT030763 AAF59292.2 AAM11251.1 CAB64381.1 EDV59713.1 ABLF02031445 ABLF02031456 ABLF02031459 OUUW01000001 SPP73099.1 CH480816 EDW46712.1 CM000157 EDW89143.2 GDIQ01042199 JAN52538.1 AGB93293.1 CH902619 EDV35887.2 AGB93294.1 EDW01588.1 CM002911 KMY91919.1 GDIP01175122 JAJ48280.1 EDV59728.2 GDIP01175123 JAJ48279.1 KMY91925.1 CH940667 EDW57330.2 GDIQ01056544 JAN38193.1 GDIQ01173399 JAK78326.1 GAMC01010064 JAB96491.1 KMY91924.1 GDIQ01056543 JAN38194.1 GDIQ01056542 JAN38195.1 GDIQ01245332 JAK06393.1 EDW31472.1 GDIQ01245331 GDIQ01245330 JAK06395.1 SPP73107.1 GBXI01005057 JAD09235.1 GDIQ01173400 JAK78325.1 KRT01536.1 GDHF01033265 JAI19049.1
NWSH01000411 PCG76591.1 KZ149965 PZC76207.1 BABH01035978 BABH01035979 BABH01035980 KQ459449 KPJ00894.1 GEBQ01005994 JAT33983.1 GECZ01002612 JAS67157.1 GEDC01011388 JAS25910.1 KQ971354 EFA06885.1 KB631648 ERL85005.1 UFQS01000067 UFQT01000067 SSW98994.1 SSX19376.1 SSW98992.1 SSX19374.1 SSW98993.1 SSX19375.1 GEZM01005749 GEZM01005748 JAV95942.1 ADMH02000160 ETN67532.1 GGFK01006475 MBW39796.1 GGFK01006379 MBW39700.1 JXJN01005875 JXJN01005876 JXJN01005877 JXJN01005878 JXJN01005879 GGFK01005759 MBW39080.1 GGFK01005868 MBW39189.1 GGFK01005772 MBW39093.1 AXCN02000591 GGMR01002668 MBY15287.1 AXCM01000153 GAKP01023256 JAC35700.1 CM000362 EDX05987.1 CVRI01000043 CRK96002.1 AAAB01008807 APCN01000218 JTDY01000684 KOB76236.1 IACF01001157 LAB66873.1 KK856713 PTY26312.1 GL732533 EFX84948.1 GBXI01007033 JAD07259.1 CH479181 EDW31467.1 GGMR01000922 MBY13541.1 GGMS01011264 MBY80467.1 CH916367 EDW01573.1 GDHF01032921 JAI19393.1 CH963849 EDW74165.1 CM000071 EAL24766.1 AJ243708 AJ243710 AJ243709 AE013599 AAF59281.2 CAB57345.3 GFXV01002542 MBW14347.1 BT126321 AEB66928.1 CH954177 KQS70970.1 EDX05972.1 AAM68907.1 AJ251892 AY094898 BT030763 AAF59292.2 AAM11251.1 CAB64381.1 EDV59713.1 ABLF02031445 ABLF02031456 ABLF02031459 OUUW01000001 SPP73099.1 CH480816 EDW46712.1 CM000157 EDW89143.2 GDIQ01042199 JAN52538.1 AGB93293.1 CH902619 EDV35887.2 AGB93294.1 EDW01588.1 CM002911 KMY91919.1 GDIP01175122 JAJ48280.1 EDV59728.2 GDIP01175123 JAJ48279.1 KMY91925.1 CH940667 EDW57330.2 GDIQ01056544 JAN38193.1 GDIQ01173399 JAK78326.1 GAMC01010064 JAB96491.1 KMY91924.1 GDIQ01056543 JAN38194.1 GDIQ01056542 JAN38195.1 GDIQ01245332 JAK06393.1 EDW31472.1 GDIQ01245331 GDIQ01245330 JAK06395.1 SPP73107.1 GBXI01005057 JAD09235.1 GDIQ01173400 JAK78325.1 KRT01536.1 GDHF01033265 JAI19049.1
Proteomes
UP000007151
UP000283053
UP000218220
UP000005204
UP000053268
UP000192223
+ More
UP000007266 UP000030742 UP000000673 UP000069272 UP000075901 UP000092460 UP000075920 UP000075886 UP000075883 UP000075884 UP000075885 UP000075900 UP000091820 UP000000304 UP000076408 UP000092445 UP000183832 UP000076407 UP000007062 UP000075840 UP000075880 UP000075882 UP000037510 UP000000305 UP000008744 UP000001070 UP000007798 UP000095301 UP000001819 UP000000803 UP000008711 UP000007819 UP000268350 UP000001292 UP000002282 UP000007801 UP000192221 UP000008792
UP000007266 UP000030742 UP000000673 UP000069272 UP000075901 UP000092460 UP000075920 UP000075886 UP000075883 UP000075884 UP000075885 UP000075900 UP000091820 UP000000304 UP000076408 UP000092445 UP000183832 UP000076407 UP000007062 UP000075840 UP000075880 UP000075882 UP000037510 UP000000305 UP000008744 UP000001070 UP000007798 UP000095301 UP000001819 UP000000803 UP000008711 UP000007819 UP000268350 UP000001292 UP000002282 UP000007801 UP000192221 UP000008792
Interpro
SUPFAM
SSF53335
SSF53335
Gene 3D
CDD
ProteinModelPortal
A0A2H1V5P8
A0A212F2G1
A0A3S2NPD8
A0A2A4JX68
A0A2W1BSM4
H9JSV8
+ More
A0A0N0P9S6 A0A1B6MDH1 A0A1B6GXL9 A0A1W4WID4 A0A1B6DJX5 D6WPW4 U4U4V9 A0A336K203 A0A336LMA4 A0A336K8B4 A0A1Y1NDG2 W5JT26 A0A182FIB0 A0A2M4AG89 A0A2M4AFY9 A0A182TBX7 A0A1B0AYM5 A0A2M4AEA4 A0A2M4AEI1 A0A2M4AE79 A0A182WIT6 A0A182QDT9 A0A2S2NDT6 A0A182MN04 A0A182NHP1 A0A182P9V8 A0A182RFQ4 A0A1A9W160 A0A034V1S2 B4QE78 A0A182XZH5 A0A1B0AAU4 A0A1J1I8E1 A0A182X6G2 Q7QJT4 A0A182I3R6 A0A182JEV1 A0A182KRW2 A0A0L7LM11 A0A2P2HYN0 A0A2R7X3E7 E9G6K1 A0A0A1X7J1 B4GCL9 A0A2S2N8J4 A0A2S2QRT3 B4J3Z0 A0A0K8TYQ3 B4MPX6 A0A1I8MQV6 Q292U5 A1Z6W3-2 A0A2H8TJM4 F3YDG0 A0A0Q5WPJ4 B4QDW0 E2QC70 Q9U1I1 B3NAF9 A1Z6W3 A1Z6W3-3 J9JLZ2 A0A3B0JI68 B4HQQ2 B4P1E1 A0A0P6G1G6 A0A0B4KED7 B3MHQ1 A0A0B4KEK5 B4J405 A0A0J9R8C4 A0A0P5C5Q7 A0A1W4V1C8 B3NAH4 A0A0P5CI28 A0A0J9TV34 B4MFG1 A0A0P6F0X0 A0A0P5LCT1 W8BTE7 A0A0J9R8C8 A0A0P6FNR5 A0A0P6G6B4 A0A0P5G7V5 A0A1W4V210 B4GCM4 A0A0P5G751 A0A3B0IYY3 A0A0A1XE64 A0A1W4VF77 A0A0P5LHW2 A0A0R3NRM5 A0A0K8TX40
A0A0N0P9S6 A0A1B6MDH1 A0A1B6GXL9 A0A1W4WID4 A0A1B6DJX5 D6WPW4 U4U4V9 A0A336K203 A0A336LMA4 A0A336K8B4 A0A1Y1NDG2 W5JT26 A0A182FIB0 A0A2M4AG89 A0A2M4AFY9 A0A182TBX7 A0A1B0AYM5 A0A2M4AEA4 A0A2M4AEI1 A0A2M4AE79 A0A182WIT6 A0A182QDT9 A0A2S2NDT6 A0A182MN04 A0A182NHP1 A0A182P9V8 A0A182RFQ4 A0A1A9W160 A0A034V1S2 B4QE78 A0A182XZH5 A0A1B0AAU4 A0A1J1I8E1 A0A182X6G2 Q7QJT4 A0A182I3R6 A0A182JEV1 A0A182KRW2 A0A0L7LM11 A0A2P2HYN0 A0A2R7X3E7 E9G6K1 A0A0A1X7J1 B4GCL9 A0A2S2N8J4 A0A2S2QRT3 B4J3Z0 A0A0K8TYQ3 B4MPX6 A0A1I8MQV6 Q292U5 A1Z6W3-2 A0A2H8TJM4 F3YDG0 A0A0Q5WPJ4 B4QDW0 E2QC70 Q9U1I1 B3NAF9 A1Z6W3 A1Z6W3-3 J9JLZ2 A0A3B0JI68 B4HQQ2 B4P1E1 A0A0P6G1G6 A0A0B4KED7 B3MHQ1 A0A0B4KEK5 B4J405 A0A0J9R8C4 A0A0P5C5Q7 A0A1W4V1C8 B3NAH4 A0A0P5CI28 A0A0J9TV34 B4MFG1 A0A0P6F0X0 A0A0P5LCT1 W8BTE7 A0A0J9R8C8 A0A0P6FNR5 A0A0P6G6B4 A0A0P5G7V5 A0A1W4V210 B4GCM4 A0A0P5G751 A0A3B0IYY3 A0A0A1XE64 A0A1W4VF77 A0A0P5LHW2 A0A0R3NRM5 A0A0K8TX40
Ontologies
GO
GO:0008270
GO:0016020
GO:0005044
GO:0004222
GO:0005886
GO:0001736
GO:0007164
GO:0009948
GO:0005737
GO:0005829
GO:0045185
GO:0001738
GO:0007163
GO:0001737
GO:0060071
GO:0030951
GO:0045773
GO:0030424
GO:0098930
GO:0045184
GO:0042067
GO:1902669
GO:0070593
GO:0050660
GO:0016491
GO:0004190
GO:0006508
GO:0005515
GO:0006811
GO:0005622
GO:0006810
GO:0003824
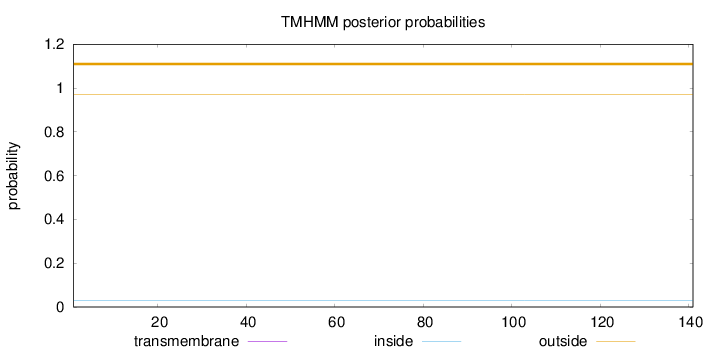
Topology
Subcellular location
Cell membrane
Length:
141
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02884
outside
1 - 141
Population Genetic Test Statistics
Pi
184.595971
Theta
189.321202
Tajima's D
0.279651
CLR
1.916109
CSRT
0.449627518624069
Interpretation
Uncertain
