Gene
KWMTBOMO11245
Pre Gene Modal
BGIBMGA012618
Annotation
PREDICTED:_glucose_dehydrogenase_[FAD?_quinone]-like_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.007 Mitochondrial Reliability : 1.465
Sequence
CDS
ATGGATTTTGCTAAATATATTCCACTTTTTGAACAATATTGCCGGTACAAAGGCGACGTTCAGTTGAAGCTCTGCGTCTCAACTCTCGGGTTGCTTTCAAAACTCCTTCTCCTTCTCACATTAACATCAGTAAAAGCTGATCTTTTCACCAAAGCGACGAGGACAAAAGCCGATGAAAGGGATCTTAGACAAAGTGTTTTTGATTTCATAATTGTCGGCGGAGGCACTGCTGGCTGCGTCTTGGCTAACAGGCTGTCAGCTGTCTCGGATTGGAAGGTTCTTTTAGTAGAAACTGGTGATGATGAGAACATAGATTATAATATCCCAGGAGTTCCGACCAATGAGTACCGTCCGATTCTGTGGAACTATAGGACAGAGAGAAACGGCTACTCCTGTCTATCCAGGCCTAGTGGAAGCTGCGAGGTCAAAACTGGAAAAGTACTCGGGGGCTCCAGTGTAACCAACGATATGAAGTATACAAGAGGCTCTAAAGTGGACTATGATGGCTGGCACAATACTGGAACGCTTGCCTCTTTGGAGTGGAAGTTCGAAAATTTAATTGATGACTTTAAATACAGCGAAGATAACGGAGATTACGACATCTTAATGAACTCATTATATCACTCTCGCGGTGGAGAGATGCACGTCAAAAAGTTCAAGTATACAGATCGATACATCGAAGTATTTAATGGAGCTTTCTCTGAAATGGGCTTACGGACCTTGGATATAAATACAAATATTCCAGAAGCTGTTGTTAATAATCAATTCGCAATGATAAACAATACTAGACTTAGTACTAATAGTGCCTTTTTGAAGCCAGTAAGACACAGGAAGAATCTAACAATTCTGAAGGGAGCTACAGTTACCAAAATTGTTATCAATAAACAAGAAAATACAGTTGATGGAATTTTGTATGAGATAGAAAAGGGTAAAGAAAGACCCGCTTATGTTAAAAGAGAAATAATTCTCACTACTGGAGCCATCAACAATGTGCGTTTGTTACACTTATCTGGAATAGGTCCAAAGATAGAATTGAAAAAGTATAATATTACTACTATCAAAGATCTTCCTGTAGGAATGAATTACCAAGATCAAGTCACCACTGGGGGTCTGGCTTTCGTGATGAACGATGTCGCACCGGTTACTGAGGAAAAAATTACTAATGATTTCAAGACCTGGTTCCAGAGCCGTAGTGGTCCGTTAGCTTCTAGAGGAATTAATCAAGTATCTGCATTTTTACAATTATATGAAAACATTAATGCTCCGGACGTTGAATTGGCATTAGATGGAAACTATATTAGAAGCGATAAGTATTTACTAACCAATCTTACCGTTCACAGCGCTGAAGAATTTAATCTACCGCTTCCTTATTATAATTTAGTTAATATTCACCCAATTTTACTGAGACCCAAAAGCAAAGGTCGAGTACTTTTAAACAAAAGAAACCCAAAGTATGGTCGACCAGTCATACAAGCAAACTTGCTAAAGGAAACAGAGGATCTGGACACTTTGGTTGATAGCATAGACATTGCTTTGCAACTCCTTGATACCAAAGAAATGAAGAAAGCAGAAGTGAAATTTGCTCCTTTAGACTTATTTCCGTGCGATAGACTAAGCAACAGAGATCAATGGAACTGCATAGCCAGACATTATACTAGAGCCATGAGTAGGCCTATTGGGACTTGTAGAATGGGAGATAATAGCACGGAGTCCGTAGTAGACTTCCAGCTTAGGGTACACGGCATCGAAGGTTTGCGCATAATTGATGCTTCTGTTATGCCTTCACACGTCCGTGGAGGCATATTTGCACCGACATTGATGATAGCAGAGAAAGGTGCAAAAATGATTTTAAAAGATTGGAAAGAAAATAAAGCTATCATTATCACAGGAGCAGCAGTTGAACCTCTAGTTATCTTAGGAATTATTGACGCATCTACCACTCTTAAATTAGAAATACCGTACACCCCCAGACTACTGTCGACAACTGATTTCGAATCAGTGACTGTACCCATCTTACATGTGCCTACGTAA
Protein
MDFAKYIPLFEQYCRYKGDVQLKLCVSTLGLLSKLLLLLTLTSVKADLFTKATRTKADERDLRQSVFDFIIVGGGTAGCVLANRLSAVSDWKVLLVETGDDENIDYNIPGVPTNEYRPILWNYRTERNGYSCLSRPSGSCEVKTGKVLGGSSVTNDMKYTRGSKVDYDGWHNTGTLASLEWKFENLIDDFKYSEDNGDYDILMNSLYHSRGGEMHVKKFKYTDRYIEVFNGAFSEMGLRTLDINTNIPEAVVNNQFAMINNTRLSTNSAFLKPVRHRKNLTILKGATVTKIVINKQENTVDGILYEIEKGKERPAYVKREIILTTGAINNVRLLHLSGIGPKIELKKYNITTIKDLPVGMNYQDQVTTGGLAFVMNDVAPVTEEKITNDFKTWFQSRSGPLASRGINQVSAFLQLYENINAPDVELALDGNYIRSDKYLLTNLTVHSAEEFNLPLPYYNLVNIHPILLRPKSKGRVLLNKRNPKYGRPVIQANLLKETEDLDTLVDSIDIALQLLDTKEMKKAEVKFAPLDLFPCDRLSNRDQWNCIARHYTRAMSRPIGTCRMGDNSTESVVDFQLRVHGIEGLRIIDASVMPSHVRGGIFAPTLMIAEKGAKMILKDWKENKAIIITGAAVEPLVILGIIDASTTLKLEIPYTPRLLSTTDFESVTVPILHVPT
Summary
Cofactor
FAD
Similarity
Belongs to the GMC oxidoreductase family.
Uniprot
H9JSV6
A0A2Z5UWZ3
A0A2W1BRG2
A0A2H1WDI9
A0A0N1INC3
A0A0N1I9N2
+ More
A0A212F2G7 A0A0L7KXH2 J9JJZ7 A0A2S2PRI8 A0A2S2QWK2 A0A2P8ZKR6 E0VFX6 K7J278 A0A154PBV5 A0A232FLV0 A0A3L8D6T0 A0A088AJU8 A0A0M8ZT63 A0A0M8ZRW9 A0A2J7RBH7 A0A0L7RJT5 K7J043 E2BJK2 A0A232EYA8 A0A310ST84 A0A0N0BBJ4 A0A2A3EHZ9 A0A067RC53 A0A2J7QFI3 A0A232EL32 A0A2J7QFJ6 A0A2J7QFH1 H9JW74 K7J0Y3 K7IPD7 A0A139WEV9 H9JWN7 A0A2A4JMY6 K7J0D1 A0A2H1X0I6 A0A2W1BFE7 A0A067R493 E0VUA7 A0A0C9QE18 A0A232FF98 A0A2P8XFT3 A0A2A4JID4 K7J281 K7J0C4 A0A224XLN0 A0A2J7Q2J7 A0A2J7Q2I0 A0A194QP52 A0A1B6D9F6 T1HCA7 A0A2J7QTE5 A0A2P8XR67 A0A232FKY7 A0A2J7RBH2 A0A2J7PRD5 K7IV19 A0A067RKI1 A0A212FML5 A0A194RET1 A0A232F8Y5 A0A0N1PFX8 A0A2S2R240 A0A026WCZ8 A0A3L8D7W0 A0A1A9WC57 K7IXL5 A0A0N0PAP0 A0A067QSX6 A0A151WMQ2 A0A2J7PES5 A0A2A4JIB7 A0A232FM80 A0A151I237 A0A194PY98 E9IKF8 A0A291P0Y5 A0A0N0P9F7 A0A1B6F402 A0A291P0N5 A0A2P8YJ25 F5HJA1 A0A195E3M0 E9IKG5 A0A2Z5UWZ1 A0A088AJU4 A0A232F996 F5HJA2 F4WAD9 A0A2J7PET3 A0A1B6DKE4 A0A1W4XEG1 A0A1B6DYE3 D2A3N1
A0A212F2G7 A0A0L7KXH2 J9JJZ7 A0A2S2PRI8 A0A2S2QWK2 A0A2P8ZKR6 E0VFX6 K7J278 A0A154PBV5 A0A232FLV0 A0A3L8D6T0 A0A088AJU8 A0A0M8ZT63 A0A0M8ZRW9 A0A2J7RBH7 A0A0L7RJT5 K7J043 E2BJK2 A0A232EYA8 A0A310ST84 A0A0N0BBJ4 A0A2A3EHZ9 A0A067RC53 A0A2J7QFI3 A0A232EL32 A0A2J7QFJ6 A0A2J7QFH1 H9JW74 K7J0Y3 K7IPD7 A0A139WEV9 H9JWN7 A0A2A4JMY6 K7J0D1 A0A2H1X0I6 A0A2W1BFE7 A0A067R493 E0VUA7 A0A0C9QE18 A0A232FF98 A0A2P8XFT3 A0A2A4JID4 K7J281 K7J0C4 A0A224XLN0 A0A2J7Q2J7 A0A2J7Q2I0 A0A194QP52 A0A1B6D9F6 T1HCA7 A0A2J7QTE5 A0A2P8XR67 A0A232FKY7 A0A2J7RBH2 A0A2J7PRD5 K7IV19 A0A067RKI1 A0A212FML5 A0A194RET1 A0A232F8Y5 A0A0N1PFX8 A0A2S2R240 A0A026WCZ8 A0A3L8D7W0 A0A1A9WC57 K7IXL5 A0A0N0PAP0 A0A067QSX6 A0A151WMQ2 A0A2J7PES5 A0A2A4JIB7 A0A232FM80 A0A151I237 A0A194PY98 E9IKF8 A0A291P0Y5 A0A0N0P9F7 A0A1B6F402 A0A291P0N5 A0A2P8YJ25 F5HJA1 A0A195E3M0 E9IKG5 A0A2Z5UWZ1 A0A088AJU4 A0A232F996 F5HJA2 F4WAD9 A0A2J7PET3 A0A1B6DKE4 A0A1W4XEG1 A0A1B6DYE3 D2A3N1
Pubmed
EMBL
BABH01035963
LC085606
BBB15976.1
KZ149965
PZC76205.1
ODYU01007937
+ More
SOQ51145.1 KQ461184 KPJ07515.1 KQ459449 KPJ00897.1 AGBW02010753 OWR47911.1 JTDY01004802 KOB67734.1 ABLF02037931 GGMR01019368 MBY31987.1 GGMS01012934 MBY82137.1 PYGN01000027 PSN57091.1 DS235129 EEB12282.1 AAZX01006810 AAZX01008181 KQ434869 KZC09322.1 NNAY01000068 OXU31327.1 QOIP01000012 RLU16177.1 KQ435889 KOX69466.1 KQ435923 KOX68393.1 NEVH01005904 PNF38171.1 KQ414579 KOC71130.1 GL448571 EFN84159.1 NNAY01001663 OXU23279.1 KQ759784 OAD62860.1 KQ435991 KOX67668.1 KZ288238 PBC31350.1 KK852802 KDR16287.1 NEVH01015297 PNF27347.1 NNAY01003669 OXU19031.1 PNF27348.1 PNF27345.1 BABH01025443 KQ971354 KYB26474.1 BABH01018352 NWSH01000973 PCG73341.1 ODYU01012451 SOQ58762.1 KZ150184 PZC72475.1 KK852991 KDR12780.1 DS235784 EEB16963.1 GBYB01012653 JAG82420.1 NNAY01000290 OXU29434.1 PYGN01002308 PSN30832.1 NWSH01001458 PCG71172.1 AAZX01008029 GFTR01007495 JAW08931.1 NEVH01019370 PNF22797.1 PNF22796.1 KQ458756 KPJ05261.1 GEDC01015002 GEDC01002775 JAS22296.1 JAS34523.1 ACPB03007733 ACPB03007734 ACPB03007735 ACPB03007736 NEVH01011195 PNF31843.1 PYGN01001493 PSN34475.1 OXU31332.1 PNF38172.1 NEVH01022362 PNF18879.1 AAZX01001198 KK852622 KDR19978.1 AGBW02007651 OWR55004.1 KQ460323 KPJ15969.1 NNAY01000675 OXU27062.1 KQ461158 KPJ08048.1 GGMS01014842 MBY84045.1 KK107261 EZA53937.1 RLU16172.1 AAZX01005632 KPJ07516.1 KK853319 KDR08567.1 KQ982937 KYQ49100.1 NEVH01026088 PNF14837.1 PCG71173.1 NNAY01000026 OXU31834.1 KQ976551 KYM80868.1 KQ459588 KPI97724.1 GL763984 EFZ18947.1 MF687602 ATJ44528.1 KPJ00896.1 GECZ01024829 JAS44940.1 MF687547 ATJ44473.1 PYGN01000560 PSN44247.1 AAAB01008844 EGK96362.1 KQ979701 KYN19666.1 EFZ18935.1 BBB15977.1 NNAY01000704 OXU26887.1 EGK96363.1 GL888048 EGI68822.1 PNF14838.1 GEDC01024026 GEDC01011147 JAS13272.1 JAS26151.1 GEDC01006623 JAS30675.1 KQ971348 EFA05531.1
SOQ51145.1 KQ461184 KPJ07515.1 KQ459449 KPJ00897.1 AGBW02010753 OWR47911.1 JTDY01004802 KOB67734.1 ABLF02037931 GGMR01019368 MBY31987.1 GGMS01012934 MBY82137.1 PYGN01000027 PSN57091.1 DS235129 EEB12282.1 AAZX01006810 AAZX01008181 KQ434869 KZC09322.1 NNAY01000068 OXU31327.1 QOIP01000012 RLU16177.1 KQ435889 KOX69466.1 KQ435923 KOX68393.1 NEVH01005904 PNF38171.1 KQ414579 KOC71130.1 GL448571 EFN84159.1 NNAY01001663 OXU23279.1 KQ759784 OAD62860.1 KQ435991 KOX67668.1 KZ288238 PBC31350.1 KK852802 KDR16287.1 NEVH01015297 PNF27347.1 NNAY01003669 OXU19031.1 PNF27348.1 PNF27345.1 BABH01025443 KQ971354 KYB26474.1 BABH01018352 NWSH01000973 PCG73341.1 ODYU01012451 SOQ58762.1 KZ150184 PZC72475.1 KK852991 KDR12780.1 DS235784 EEB16963.1 GBYB01012653 JAG82420.1 NNAY01000290 OXU29434.1 PYGN01002308 PSN30832.1 NWSH01001458 PCG71172.1 AAZX01008029 GFTR01007495 JAW08931.1 NEVH01019370 PNF22797.1 PNF22796.1 KQ458756 KPJ05261.1 GEDC01015002 GEDC01002775 JAS22296.1 JAS34523.1 ACPB03007733 ACPB03007734 ACPB03007735 ACPB03007736 NEVH01011195 PNF31843.1 PYGN01001493 PSN34475.1 OXU31332.1 PNF38172.1 NEVH01022362 PNF18879.1 AAZX01001198 KK852622 KDR19978.1 AGBW02007651 OWR55004.1 KQ460323 KPJ15969.1 NNAY01000675 OXU27062.1 KQ461158 KPJ08048.1 GGMS01014842 MBY84045.1 KK107261 EZA53937.1 RLU16172.1 AAZX01005632 KPJ07516.1 KK853319 KDR08567.1 KQ982937 KYQ49100.1 NEVH01026088 PNF14837.1 PCG71173.1 NNAY01000026 OXU31834.1 KQ976551 KYM80868.1 KQ459588 KPI97724.1 GL763984 EFZ18947.1 MF687602 ATJ44528.1 KPJ00896.1 GECZ01024829 JAS44940.1 MF687547 ATJ44473.1 PYGN01000560 PSN44247.1 AAAB01008844 EGK96362.1 KQ979701 KYN19666.1 EFZ18935.1 BBB15977.1 NNAY01000704 OXU26887.1 EGK96363.1 GL888048 EGI68822.1 PNF14838.1 GEDC01024026 GEDC01011147 JAS13272.1 JAS26151.1 GEDC01006623 JAS30675.1 KQ971348 EFA05531.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000037510
UP000007819
+ More
UP000245037 UP000009046 UP000002358 UP000076502 UP000215335 UP000279307 UP000005203 UP000053105 UP000235965 UP000053825 UP000008237 UP000242457 UP000027135 UP000007266 UP000218220 UP000015103 UP000053097 UP000091820 UP000075809 UP000078540 UP000007062 UP000078492 UP000007755 UP000192223
UP000245037 UP000009046 UP000002358 UP000076502 UP000215335 UP000279307 UP000005203 UP000053105 UP000235965 UP000053825 UP000008237 UP000242457 UP000027135 UP000007266 UP000218220 UP000015103 UP000053097 UP000091820 UP000075809 UP000078540 UP000007062 UP000078492 UP000007755 UP000192223
Interpro
SUPFAM
SSF51905
SSF51905
Gene 3D
CDD
ProteinModelPortal
H9JSV6
A0A2Z5UWZ3
A0A2W1BRG2
A0A2H1WDI9
A0A0N1INC3
A0A0N1I9N2
+ More
A0A212F2G7 A0A0L7KXH2 J9JJZ7 A0A2S2PRI8 A0A2S2QWK2 A0A2P8ZKR6 E0VFX6 K7J278 A0A154PBV5 A0A232FLV0 A0A3L8D6T0 A0A088AJU8 A0A0M8ZT63 A0A0M8ZRW9 A0A2J7RBH7 A0A0L7RJT5 K7J043 E2BJK2 A0A232EYA8 A0A310ST84 A0A0N0BBJ4 A0A2A3EHZ9 A0A067RC53 A0A2J7QFI3 A0A232EL32 A0A2J7QFJ6 A0A2J7QFH1 H9JW74 K7J0Y3 K7IPD7 A0A139WEV9 H9JWN7 A0A2A4JMY6 K7J0D1 A0A2H1X0I6 A0A2W1BFE7 A0A067R493 E0VUA7 A0A0C9QE18 A0A232FF98 A0A2P8XFT3 A0A2A4JID4 K7J281 K7J0C4 A0A224XLN0 A0A2J7Q2J7 A0A2J7Q2I0 A0A194QP52 A0A1B6D9F6 T1HCA7 A0A2J7QTE5 A0A2P8XR67 A0A232FKY7 A0A2J7RBH2 A0A2J7PRD5 K7IV19 A0A067RKI1 A0A212FML5 A0A194RET1 A0A232F8Y5 A0A0N1PFX8 A0A2S2R240 A0A026WCZ8 A0A3L8D7W0 A0A1A9WC57 K7IXL5 A0A0N0PAP0 A0A067QSX6 A0A151WMQ2 A0A2J7PES5 A0A2A4JIB7 A0A232FM80 A0A151I237 A0A194PY98 E9IKF8 A0A291P0Y5 A0A0N0P9F7 A0A1B6F402 A0A291P0N5 A0A2P8YJ25 F5HJA1 A0A195E3M0 E9IKG5 A0A2Z5UWZ1 A0A088AJU4 A0A232F996 F5HJA2 F4WAD9 A0A2J7PET3 A0A1B6DKE4 A0A1W4XEG1 A0A1B6DYE3 D2A3N1
A0A212F2G7 A0A0L7KXH2 J9JJZ7 A0A2S2PRI8 A0A2S2QWK2 A0A2P8ZKR6 E0VFX6 K7J278 A0A154PBV5 A0A232FLV0 A0A3L8D6T0 A0A088AJU8 A0A0M8ZT63 A0A0M8ZRW9 A0A2J7RBH7 A0A0L7RJT5 K7J043 E2BJK2 A0A232EYA8 A0A310ST84 A0A0N0BBJ4 A0A2A3EHZ9 A0A067RC53 A0A2J7QFI3 A0A232EL32 A0A2J7QFJ6 A0A2J7QFH1 H9JW74 K7J0Y3 K7IPD7 A0A139WEV9 H9JWN7 A0A2A4JMY6 K7J0D1 A0A2H1X0I6 A0A2W1BFE7 A0A067R493 E0VUA7 A0A0C9QE18 A0A232FF98 A0A2P8XFT3 A0A2A4JID4 K7J281 K7J0C4 A0A224XLN0 A0A2J7Q2J7 A0A2J7Q2I0 A0A194QP52 A0A1B6D9F6 T1HCA7 A0A2J7QTE5 A0A2P8XR67 A0A232FKY7 A0A2J7RBH2 A0A2J7PRD5 K7IV19 A0A067RKI1 A0A212FML5 A0A194RET1 A0A232F8Y5 A0A0N1PFX8 A0A2S2R240 A0A026WCZ8 A0A3L8D7W0 A0A1A9WC57 K7IXL5 A0A0N0PAP0 A0A067QSX6 A0A151WMQ2 A0A2J7PES5 A0A2A4JIB7 A0A232FM80 A0A151I237 A0A194PY98 E9IKF8 A0A291P0Y5 A0A0N0P9F7 A0A1B6F402 A0A291P0N5 A0A2P8YJ25 F5HJA1 A0A195E3M0 E9IKG5 A0A2Z5UWZ1 A0A088AJU4 A0A232F996 F5HJA2 F4WAD9 A0A2J7PET3 A0A1B6DKE4 A0A1W4XEG1 A0A1B6DYE3 D2A3N1
PDB
5NCC
E-value=8.19539e-56,
Score=552
Ontologies
GO
Topology
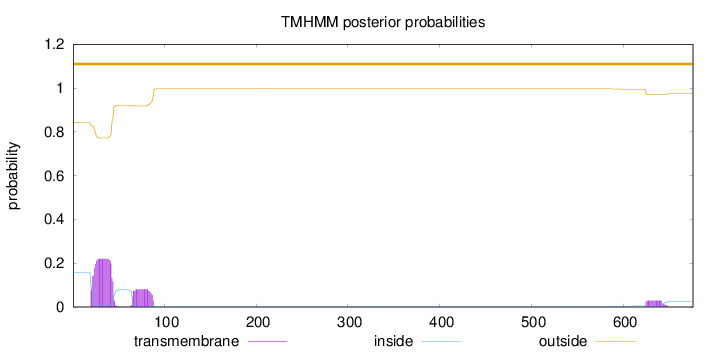
Length:
676
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
7.21800999999999
Exp number, first 60 AAs:
4.84107
Total prob of N-in:
0.15753
outside
1 - 676
Population Genetic Test Statistics
Pi
166.656645
Theta
169.741354
Tajima's D
-0.884771
CLR
1.308998
CSRT
0.160641967901605
Interpretation
Uncertain
