Gene
KWMTBOMO11240
Annotation
PREDICTED:_uncharacterized_protein_LOC106713896?_partial_[Papilio_machaon]
Location in the cell
Mitochondrial Reliability : 1.522 PlasmaMembrane Reliability : 1.124
Sequence
CDS
ATGTATCGGGATGTCGTTGCAGTCGATAAGGGGCTAACTCCGCTATCCTTAGCTAACGGGAATTGGAAGAGGCTGAAGGTAGAGGGTATTTCAGATCGCATAACCGTATGGAGGAGCAAGGAGTTACACGGTAGGTTCTACCGGGCTCTCACAGGGCCTGATGTAGACCAATTAGCTTCCGTGGCCTGGTTACGATTCAGTAACCTCTTTGGGGAAACTGAAGGTTTTATCTTTGCAATTATGGACGAAGTTATCATGACGAACAACTACCGGAAATATATACTAAAGGATGGGACGGTCGACATTTGTCGGGCATGCCACCGTCCGGGAGAGTCTATTAGACATATTGTTTCTGGCTGTTCTCGGTTTGCTAACGGCGAGTACTTGCATAGACATAACCAAGTGGCTAGGATTATCCATCAACAACTTGCTCTGAAATATGGTTTTCTTGATTCTGCTGTACCTTATTACCAATACCAACCTCAACCAGTTCTTGATAATGGTCATGTTACGCTCTACTGGGACCGATCTATTATCACGGATAGGTATATTGTTGCCAATAAACCTGATATTGTGATAATAGATCGATCTGCGCGTCGAGCAGTGTTGGTTGATGTCACCATTCCTCATGACGAGAACCTCGTGAAGGCAGAAAAGGACAAATTAATAAAATATTTAGACCTGGCCCACGAGACTACCGCCATGTGGCATGTTGATTCAACCATCATTGTTCCAATAGTTGTATCGGTACACGGCTTGATAGCGAAAAGTTTCGATCAACATCTTAAGAAACTTTCGTTATCCAGCTGGGTTAAGGGCCTAATACAGAAAGCAGTTATCCTCGATACTGCACGCATTGTGAGGAGGTTTCTCACTCTGGAGTCCTAA
Protein
MYRDVVAVDKGLTPLSLANGNWKRLKVEGISDRITVWRSKELHGRFYRALTGPDVDQLASVAWLRFSNLFGETEGFIFAIMDEVIMTNNYRKYILKDGTVDICRACHRPGESIRHIVSGCSRFANGEYLHRHNQVARIIHQQLALKYGFLDSAVPYYQYQPQPVLDNGHVTLYWDRSIITDRYIVANKPDIVIIDRSARRAVLVDVTIPHDENLVKAEKDKLIKYLDLAHETTAMWHVDSTIIVPIVVSVHGLIAKSFDQHLKKLSLSSWVKGLIQKAVILDTARIVRRFLTLES
Summary
Uniprot
A0A3S2LN57
A0A2A4JL63
A0A2H1WHZ3
A0A3S2ND61
A0A2H1VUG2
A0A2W1BC23
+ More
A0A336M9S3 A0A1Y1MG39 A0A2H1WV36 A0A1S3CYA3 A0A1Y1MCA6 A0A1Y1MT60 A0A1S3DFN1 W8BF34 H3ATI1 H3A9J0 D7GY63 H3AS73 H3AA26 A0A2H1VQZ6 A0A1S4EQ35 Q76IN5 H3B0R3 A0A1S3DB34 D7GYL6 H3AB60 W8AXW6 H2ZWF0 H3BBL0 A0A2H1VMW0 A0A3S3RHE8 A0A2J7R2D2 H3AA24 A0A2J7PIT9 E0VA19 A0A2J7PDQ6 A0A2J7RGT7 E0W0H3 E0W0M4 E0VPK6 E0VSD9 E0VPJ4 H3AHZ1 H3BFH8 A0A2J7PSM8 D7GYE3 H2ZXX1 H3B0S5 A0A3B3I0I7 A0A3P9MNJ5 R4G9X1 H3A542 H2ZUK9 H3AYE9 R4GCF3 A0A3P8NFL6 H3AKE8 A0A3P8PU69 R4GAP2 H3A541 R4GAQ0 A0A3B3HW39 A0A3B3HU40 A0A3B3HQX9 A0A2I4AQW5 A0A3P9K6Z6 H3ARW9 A0A087XGF1 A0A3P9LVD0 A0A3P8P1Q8 A0A3B3H8G1 A0A3B5PPD0 A0A3P9MMW4 A0A3B3ILW9 A0A3P9M4C8 A0A3P8NVY4 A0A3P8NT65 H3APX3 A0A3P8R5E8 A0A3B3I5C9 A0A3P9QFG6 A0A3P8NRB8 A0A3P8Q816 A0A3P8QAR5 A0A3P8PIV2 A0A3P8PST2 A0A3P8Q8N4 H3AFG1 A0A3P8PUT7 A0A3P8NLS5
A0A336M9S3 A0A1Y1MG39 A0A2H1WV36 A0A1S3CYA3 A0A1Y1MCA6 A0A1Y1MT60 A0A1S3DFN1 W8BF34 H3ATI1 H3A9J0 D7GY63 H3AS73 H3AA26 A0A2H1VQZ6 A0A1S4EQ35 Q76IN5 H3B0R3 A0A1S3DB34 D7GYL6 H3AB60 W8AXW6 H2ZWF0 H3BBL0 A0A2H1VMW0 A0A3S3RHE8 A0A2J7R2D2 H3AA24 A0A2J7PIT9 E0VA19 A0A2J7PDQ6 A0A2J7RGT7 E0W0H3 E0W0M4 E0VPK6 E0VSD9 E0VPJ4 H3AHZ1 H3BFH8 A0A2J7PSM8 D7GYE3 H2ZXX1 H3B0S5 A0A3B3I0I7 A0A3P9MNJ5 R4G9X1 H3A542 H2ZUK9 H3AYE9 R4GCF3 A0A3P8NFL6 H3AKE8 A0A3P8PU69 R4GAP2 H3A541 R4GAQ0 A0A3B3HW39 A0A3B3HU40 A0A3B3HQX9 A0A2I4AQW5 A0A3P9K6Z6 H3ARW9 A0A087XGF1 A0A3P9LVD0 A0A3P8P1Q8 A0A3B3H8G1 A0A3B5PPD0 A0A3P9MMW4 A0A3B3ILW9 A0A3P9M4C8 A0A3P8NVY4 A0A3P8NT65 H3APX3 A0A3P8R5E8 A0A3B3I5C9 A0A3P9QFG6 A0A3P8NRB8 A0A3P8Q816 A0A3P8QAR5 A0A3P8PIV2 A0A3P8PST2 A0A3P8Q8N4 H3AFG1 A0A3P8PUT7 A0A3P8NLS5
Pubmed
EMBL
RSAL01001347
RVE40825.1
NWSH01001199
PCG72152.1
ODYU01008727
SOQ52576.1
+ More
RSAL01000200 RVE44451.1 ODYU01004516 SOQ44481.1 KZ150607 PZC70460.1 UFQT01000728 SSX26770.1 GEZM01035222 JAV83335.1 ODYU01011284 SOQ56923.1 GEZM01035223 JAV83334.1 GEZM01022180 JAV88769.1 GAMC01018221 JAB88334.1 AFYH01046062 AFYH01147180 GG695107 EFA13287.1 AFYH01158743 AFYH01207636 ODYU01003901 SOQ43240.1 AB097127 BAC82595.1 AFYH01017561 GG695857 EFA13462.1 AFYH01227415 GAMC01016897 JAB89658.1 AFYH01259886 AFYH01052121 ODYU01003427 SOQ42175.1 NCKU01013699 RWR99758.1 NEVH01007832 PNF34997.1 NEVH01024955 PNF16260.1 DS235004 EEB10225.1 NEVH01026385 PNF14463.1 NEVH01003752 PNF40040.1 DS235859 EEB19129.1 DS235861 EEB19180.1 DS235366 EEB15312.1 DS235750 EEB16295.1 EEB15300.1 AFYH01167468 AFYH01026690 NEVH01021925 PNF19338.1 GG695465 EFA13373.1 AFYH01264165 AFYH01092381 AFYH01092382 AAWZ02024166 AFYH01248595 AFYH01268965 AFYH01132769 AAWZ02035408 AFYH01140129 AAWZ02036698 AFYH01154158 AYCK01026151 AFYH01155094 AFYH01154199
RSAL01000200 RVE44451.1 ODYU01004516 SOQ44481.1 KZ150607 PZC70460.1 UFQT01000728 SSX26770.1 GEZM01035222 JAV83335.1 ODYU01011284 SOQ56923.1 GEZM01035223 JAV83334.1 GEZM01022180 JAV88769.1 GAMC01018221 JAB88334.1 AFYH01046062 AFYH01147180 GG695107 EFA13287.1 AFYH01158743 AFYH01207636 ODYU01003901 SOQ43240.1 AB097127 BAC82595.1 AFYH01017561 GG695857 EFA13462.1 AFYH01227415 GAMC01016897 JAB89658.1 AFYH01259886 AFYH01052121 ODYU01003427 SOQ42175.1 NCKU01013699 RWR99758.1 NEVH01007832 PNF34997.1 NEVH01024955 PNF16260.1 DS235004 EEB10225.1 NEVH01026385 PNF14463.1 NEVH01003752 PNF40040.1 DS235859 EEB19129.1 DS235861 EEB19180.1 DS235366 EEB15312.1 DS235750 EEB16295.1 EEB15300.1 AFYH01167468 AFYH01026690 NEVH01021925 PNF19338.1 GG695465 EFA13373.1 AFYH01264165 AFYH01092381 AFYH01092382 AAWZ02024166 AFYH01248595 AFYH01268965 AFYH01132769 AAWZ02035408 AFYH01140129 AAWZ02036698 AFYH01154158 AYCK01026151 AFYH01155094 AFYH01154199
Proteomes
Pfam
PF00078 RVT_1
ProteinModelPortal
A0A3S2LN57
A0A2A4JL63
A0A2H1WHZ3
A0A3S2ND61
A0A2H1VUG2
A0A2W1BC23
+ More
A0A336M9S3 A0A1Y1MG39 A0A2H1WV36 A0A1S3CYA3 A0A1Y1MCA6 A0A1Y1MT60 A0A1S3DFN1 W8BF34 H3ATI1 H3A9J0 D7GY63 H3AS73 H3AA26 A0A2H1VQZ6 A0A1S4EQ35 Q76IN5 H3B0R3 A0A1S3DB34 D7GYL6 H3AB60 W8AXW6 H2ZWF0 H3BBL0 A0A2H1VMW0 A0A3S3RHE8 A0A2J7R2D2 H3AA24 A0A2J7PIT9 E0VA19 A0A2J7PDQ6 A0A2J7RGT7 E0W0H3 E0W0M4 E0VPK6 E0VSD9 E0VPJ4 H3AHZ1 H3BFH8 A0A2J7PSM8 D7GYE3 H2ZXX1 H3B0S5 A0A3B3I0I7 A0A3P9MNJ5 R4G9X1 H3A542 H2ZUK9 H3AYE9 R4GCF3 A0A3P8NFL6 H3AKE8 A0A3P8PU69 R4GAP2 H3A541 R4GAQ0 A0A3B3HW39 A0A3B3HU40 A0A3B3HQX9 A0A2I4AQW5 A0A3P9K6Z6 H3ARW9 A0A087XGF1 A0A3P9LVD0 A0A3P8P1Q8 A0A3B3H8G1 A0A3B5PPD0 A0A3P9MMW4 A0A3B3ILW9 A0A3P9M4C8 A0A3P8NVY4 A0A3P8NT65 H3APX3 A0A3P8R5E8 A0A3B3I5C9 A0A3P9QFG6 A0A3P8NRB8 A0A3P8Q816 A0A3P8QAR5 A0A3P8PIV2 A0A3P8PST2 A0A3P8Q8N4 H3AFG1 A0A3P8PUT7 A0A3P8NLS5
A0A336M9S3 A0A1Y1MG39 A0A2H1WV36 A0A1S3CYA3 A0A1Y1MCA6 A0A1Y1MT60 A0A1S3DFN1 W8BF34 H3ATI1 H3A9J0 D7GY63 H3AS73 H3AA26 A0A2H1VQZ6 A0A1S4EQ35 Q76IN5 H3B0R3 A0A1S3DB34 D7GYL6 H3AB60 W8AXW6 H2ZWF0 H3BBL0 A0A2H1VMW0 A0A3S3RHE8 A0A2J7R2D2 H3AA24 A0A2J7PIT9 E0VA19 A0A2J7PDQ6 A0A2J7RGT7 E0W0H3 E0W0M4 E0VPK6 E0VSD9 E0VPJ4 H3AHZ1 H3BFH8 A0A2J7PSM8 D7GYE3 H2ZXX1 H3B0S5 A0A3B3I0I7 A0A3P9MNJ5 R4G9X1 H3A542 H2ZUK9 H3AYE9 R4GCF3 A0A3P8NFL6 H3AKE8 A0A3P8PU69 R4GAP2 H3A541 R4GAQ0 A0A3B3HW39 A0A3B3HU40 A0A3B3HQX9 A0A2I4AQW5 A0A3P9K6Z6 H3ARW9 A0A087XGF1 A0A3P9LVD0 A0A3P8P1Q8 A0A3B3H8G1 A0A3B5PPD0 A0A3P9MMW4 A0A3B3ILW9 A0A3P9M4C8 A0A3P8NVY4 A0A3P8NT65 H3APX3 A0A3P8R5E8 A0A3B3I5C9 A0A3P9QFG6 A0A3P8NRB8 A0A3P8Q816 A0A3P8QAR5 A0A3P8PIV2 A0A3P8PST2 A0A3P8Q8N4 H3AFG1 A0A3P8PUT7 A0A3P8NLS5
Ontologies
GO
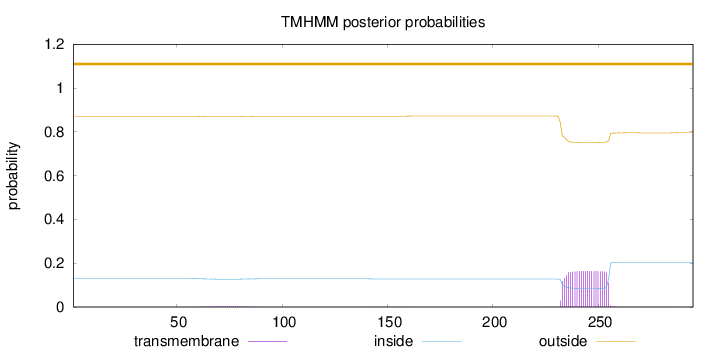
Topology
Length:
295
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.73578
Exp number, first 60 AAs:
0.00358
Total prob of N-in:
0.12976
outside
1 - 295
Population Genetic Test Statistics
Pi
152.423496
Theta
80.010538
Tajima's D
2.699988
CLR
21.372855
CSRT
0.96085195740213
Interpretation
Uncertain
