Gene
KWMTBOMO11229 Validated by peptides from experiments
Annotation
PREDICTED:_basigin_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.09
Sequence
CDS
ATGAAAGTGAAAATAGTGAAAAATATAACTATGCGATCTCAACTGTGTTTGCCTTTATCTCTTCTATTTTTAGCCTGCGGAGTTCTTACGAGTGCCCAAAACTCGTCAACGATTGCTCCGGTCACCGAGCCCACCCAAGAACTGGTCAATTATGAGGCCTTGAAGTACATCGTTGGACGCCCGTTCAATTTGAATTGCACTCTCGCCGTGCCACTGGATTCGTTTGAGATCGTATGGAAGAAGAACAGCGTGCCTCTCGGCGAGGTGGCCGGCCTCAAGGAGCGGTACGAGCTCCAGAACAAGGGGCTCACCTTCCAGATCAAGGGCCGCAGTAACGAGGACGACTACGGCAACTACACGTGCGGCCTCAAGAACCAAACGGGACATATCAAAGCCTGGATGGTCACGGGAAACGTGCACGCCAAGATGACCAAGGACGCCAACGTGGTCGAGGGACAGAACATTAAGATCACTTGCAAGTTGATCGGCAAGCCGTACTCTGAGGTGACGTGGAAGTACAAGAAGGACGAGTTGGACAACGGCACGGACGTGAGCGCGGTGCTGGGCAGTCGCGTGCAGCTGGAGAAGAACGAGCAGGGCCTGGACAACACCGTGCTGGTGCTGCAGAACGCGGAGCGCGCCGACGCCGGCCTGTACCAGTGCAGCCCGGCCGGGGGCACGCCCGCCGACGTCACGCTCAGGGTCAAGGGCGTGTACGACGCGCTCTGGCCCTTCCTCGGCATCTGCGCGGAGGTCGTGGTGCTCTGCGCCGTCATACTCTTCTACGAGAAGAGGAGGACCAAGCCCGAGCTCGACGACTCCGACACCGACAACCACGACCAGAAGAAGTCGTAG
Protein
MKVKIVKNITMRSQLCLPLSLLFLACGVLTSAQNSSTIAPVTEPTQELVNYEALKYIVGRPFNLNCTLAVPLDSFEIVWKKNSVPLGEVAGLKERYELQNKGLTFQIKGRSNEDDYGNYTCGLKNQTGHIKAWMVTGNVHAKMTKDANVVEGQNIKITCKLIGKPYSEVTWKYKKDELDNGTDVSAVLGSRVQLEKNEQGLDNTVLVLQNAERADAGLYQCSPAGGTPADVTLRVKGVYDALWPFLGICAEVVVLCAVILFYEKRRTKPELDDSDTDNHDQKKS
Summary
Uniprot
A0A2W1BRF1
A0A3B0E3L9
A0A212F2E6
A0A0N1IN03
A0A0N1PHP0
A0A0Q9WS22
+ More
A0A1B6FKJ1 B3MMX2 A0A1B6M8H5 A0A1W4WZJ0 A0A1Y1LSW9 A0A1B6LW55 T1E289 A0A0Q5WBY4 A0A0J9QXX0 U5EQY7 T1DEN1 A0A2J7R3K5 Q16ML7 T1D4G7 Q16ML6 Q961I4 A0A0J9QZ03 A0A023ENP6 Q8IPG9 A0A0J9QYA8 A0A0J9TIG8 D3TLY5 B4HYE5 M9PCW5 Q7KTJ7 A0A2M4ABL6 A0A2M4AB32 A0A182PLT5 A0A2M4DNX6 A0A2M4DMI6 A0A0J9QXT7 N6SRJ7 A0A2M4AB34 A0A2M4AAX2 A0A2P6LA76 V5GVB3 A0A0A1XRN1 A0A182XFV9 A0A0J9QYA0 T1DHN0 A0A0J9TIH2 A0A0A9W6U6 A0A2M4DMK3 A0A1L8DA04 A0A1S4GY62 A0A1B6D4E0 E0VQ49 A0A1W7RA30 A0A182F3H9 A0A1B6DMQ0 A0A2M3Z483 A0A1Q3G3R7 A0A2M4DMQ8 A0A0A9X5C1 D6WQV4 A0A1Q3FY83 A0A1Q3FYB0 A0A2M3ZE27 A0A0N8ES29 A0A1W4VWC6 A0A0R1DT06 A0A1I8NUC4 Q29L42 A0A1Q3FY55 A0A0M4E994 W8AD12 A0A2M4BVQ8 A0A2M4BWT8 A0A224YVE7 A0A0K8RFE5 R4WD95 A0A0K8UG24 Q7Q4I1 A0A336LT56 B4KEC5 L7MBI1 A0A2S2R9Q4 L7MHV3 A0A1V9XKW6 A0A2C9GPZ7 B7QIE6 A0A3L8DRF8 A0A2C9GQ66 A0A087UAU7 A0A026WUC4 A0A182MYY3 A0A2C9GQ04 A0A182L0G9 L7M0I3
A0A1B6FKJ1 B3MMX2 A0A1B6M8H5 A0A1W4WZJ0 A0A1Y1LSW9 A0A1B6LW55 T1E289 A0A0Q5WBY4 A0A0J9QXX0 U5EQY7 T1DEN1 A0A2J7R3K5 Q16ML7 T1D4G7 Q16ML6 Q961I4 A0A0J9QZ03 A0A023ENP6 Q8IPG9 A0A0J9QYA8 A0A0J9TIG8 D3TLY5 B4HYE5 M9PCW5 Q7KTJ7 A0A2M4ABL6 A0A2M4AB32 A0A182PLT5 A0A2M4DNX6 A0A2M4DMI6 A0A0J9QXT7 N6SRJ7 A0A2M4AB34 A0A2M4AAX2 A0A2P6LA76 V5GVB3 A0A0A1XRN1 A0A182XFV9 A0A0J9QYA0 T1DHN0 A0A0J9TIH2 A0A0A9W6U6 A0A2M4DMK3 A0A1L8DA04 A0A1S4GY62 A0A1B6D4E0 E0VQ49 A0A1W7RA30 A0A182F3H9 A0A1B6DMQ0 A0A2M3Z483 A0A1Q3G3R7 A0A2M4DMQ8 A0A0A9X5C1 D6WQV4 A0A1Q3FY83 A0A1Q3FYB0 A0A2M3ZE27 A0A0N8ES29 A0A1W4VWC6 A0A0R1DT06 A0A1I8NUC4 Q29L42 A0A1Q3FY55 A0A0M4E994 W8AD12 A0A2M4BVQ8 A0A2M4BWT8 A0A224YVE7 A0A0K8RFE5 R4WD95 A0A0K8UG24 Q7Q4I1 A0A336LT56 B4KEC5 L7MBI1 A0A2S2R9Q4 L7MHV3 A0A1V9XKW6 A0A2C9GPZ7 B7QIE6 A0A3L8DRF8 A0A2C9GQ66 A0A087UAU7 A0A026WUC4 A0A182MYY3 A0A2C9GQ04 A0A182L0G9 L7M0I3
Pubmed
28756777
22118469
26354079
17994087
18057021
28004739
+ More
24330624 22936249 17510324 24945155 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20353571 23537049 25830018 25401762 26823975 12364791 20566863 18362917 19820115 26999592 17550304 15632085 24495485 28797301 23691247 25576852 28327890 30249741 24508170 20966253
24330624 22936249 17510324 24945155 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20353571 23537049 25830018 25401762 26823975 12364791 20566863 18362917 19820115 26999592 17550304 15632085 24495485 28797301 23691247 25576852 28327890 30249741 24508170 20966253
EMBL
KZ149965
PZC76195.1
RBVL01000249
RKO10146.1
AGBW02010753
OWR47894.1
+ More
KQ459449 KPJ00918.1 KQ461184 KPJ07500.1 CH963857 KRF98397.1 GECZ01019256 JAS50513.1 CH902620 EDV30997.2 KPU73178.1 KPU73179.1 GEBQ01007823 JAT32154.1 GEZM01049149 JAV76071.1 GEBQ01012072 JAT27905.1 GALA01000954 JAA93898.1 CH954177 KQS70735.1 KQS70736.1 KQS70737.1 KQS70738.1 KQS70739.1 KQS70740.1 KQS70741.1 CM002910 KMY88927.1 GANO01003997 JAB55874.1 GALA01000967 JAA93885.1 NEVH01007819 PNF35400.1 CH477858 EAT35573.1 GALA01000968 JAA93884.1 EAT35571.1 EAT35572.1 EAT35574.1 AY051571 AAK92995.1 KMY88918.1 KMY88919.1 KMY88921.1 KMY88923.1 GAPW01003042 GEHC01001388 JAC10556.1 JAV46257.1 AE014134 BT005198 BT006333 AAN10653.1 AAN10654.1 AAN10655.1 AAN10656.1 AAN10657.1 AAN10658.1 AAN10659.1 AAN10660.1 AAO61755.1 AAP20829.1 KMY88928.1 KMY88920.1 KMY88922.1 KMY88924.1 EZ422437 ADD18713.1 CH480818 EDW52075.1 AGB92760.1 AAN10661.2 GGFK01004687 MBW38008.1 GGFK01004674 MBW37995.1 GGFL01014640 MBW78818.1 GGFL01014602 MBW78780.1 KMY88917.1 APGK01059412 KB741293 KB632046 ENN70239.1 ERL88250.1 GGFK01004601 MBW37922.1 GGFK01004606 MBW37927.1 MWRG01000736 PRD35473.1 GALX01004293 JAB64173.1 GBXI01000660 JAD13632.1 KMY88926.1 GAMD01002256 JAA99334.1 KMY88925.1 GBHO01041381 GBRD01012929 GDHC01008080 JAG02223.1 JAG52897.1 JAQ10549.1 GGFL01014604 MBW78782.1 GFDF01010924 JAV03160.1 AAAB01008964 GEDC01016729 JAS20569.1 DS235389 EEB15505.1 GFAH01000388 JAV48001.1 GEDC01010337 JAS26961.1 GGFM01002575 MBW23326.1 GFDL01000597 JAV34448.1 GGFL01014641 MBW78819.1 GBHO01029439 GBHO01027677 GDHC01020275 JAG14165.1 JAG15927.1 JAP98353.1 KQ971351 EFA06512.2 GFDL01002539 JAV32506.1 GFDL01002528 JAV32517.1 GGFM01006035 MBW26786.1 GDUN01000618 JAN95301.1 CM000157 KRJ97791.1 KRJ97792.1 KRJ97793.1 KRJ97794.1 KRJ97796.1 KRJ97797.1 KRJ97798.1 CH379061 EAL32982.2 GFDL01002560 JAV32485.1 CP012523 ALC38726.1 GAMC01020680 JAB85875.1 GGFJ01008016 MBW57157.1 GGFJ01008047 MBW57188.1 GFPF01008563 MAA19709.1 GADI01004007 JAA69801.1 AK417493 BAN20708.1 GDHF01026700 JAI25614.1 EAA12759.4 UFQS01000180 UFQT01000180 SSX00826.1 SSX21206.1 CH933807 EDW12893.2 GACK01004491 JAA60543.1 GGMS01017526 MBY86729.1 GACK01002275 JAA62759.1 MNPL01008544 OQR74180.1 APCN01000774 ABJB010175462 ABJB010273414 ABJB010820902 DS945857 EEC18618.1 QOIP01000005 RLU22378.1 KK119043 KFM74486.1 KK107102 EZA59607.1 GACK01008396 JAA56638.1
KQ459449 KPJ00918.1 KQ461184 KPJ07500.1 CH963857 KRF98397.1 GECZ01019256 JAS50513.1 CH902620 EDV30997.2 KPU73178.1 KPU73179.1 GEBQ01007823 JAT32154.1 GEZM01049149 JAV76071.1 GEBQ01012072 JAT27905.1 GALA01000954 JAA93898.1 CH954177 KQS70735.1 KQS70736.1 KQS70737.1 KQS70738.1 KQS70739.1 KQS70740.1 KQS70741.1 CM002910 KMY88927.1 GANO01003997 JAB55874.1 GALA01000967 JAA93885.1 NEVH01007819 PNF35400.1 CH477858 EAT35573.1 GALA01000968 JAA93884.1 EAT35571.1 EAT35572.1 EAT35574.1 AY051571 AAK92995.1 KMY88918.1 KMY88919.1 KMY88921.1 KMY88923.1 GAPW01003042 GEHC01001388 JAC10556.1 JAV46257.1 AE014134 BT005198 BT006333 AAN10653.1 AAN10654.1 AAN10655.1 AAN10656.1 AAN10657.1 AAN10658.1 AAN10659.1 AAN10660.1 AAO61755.1 AAP20829.1 KMY88928.1 KMY88920.1 KMY88922.1 KMY88924.1 EZ422437 ADD18713.1 CH480818 EDW52075.1 AGB92760.1 AAN10661.2 GGFK01004687 MBW38008.1 GGFK01004674 MBW37995.1 GGFL01014640 MBW78818.1 GGFL01014602 MBW78780.1 KMY88917.1 APGK01059412 KB741293 KB632046 ENN70239.1 ERL88250.1 GGFK01004601 MBW37922.1 GGFK01004606 MBW37927.1 MWRG01000736 PRD35473.1 GALX01004293 JAB64173.1 GBXI01000660 JAD13632.1 KMY88926.1 GAMD01002256 JAA99334.1 KMY88925.1 GBHO01041381 GBRD01012929 GDHC01008080 JAG02223.1 JAG52897.1 JAQ10549.1 GGFL01014604 MBW78782.1 GFDF01010924 JAV03160.1 AAAB01008964 GEDC01016729 JAS20569.1 DS235389 EEB15505.1 GFAH01000388 JAV48001.1 GEDC01010337 JAS26961.1 GGFM01002575 MBW23326.1 GFDL01000597 JAV34448.1 GGFL01014641 MBW78819.1 GBHO01029439 GBHO01027677 GDHC01020275 JAG14165.1 JAG15927.1 JAP98353.1 KQ971351 EFA06512.2 GFDL01002539 JAV32506.1 GFDL01002528 JAV32517.1 GGFM01006035 MBW26786.1 GDUN01000618 JAN95301.1 CM000157 KRJ97791.1 KRJ97792.1 KRJ97793.1 KRJ97794.1 KRJ97796.1 KRJ97797.1 KRJ97798.1 CH379061 EAL32982.2 GFDL01002560 JAV32485.1 CP012523 ALC38726.1 GAMC01020680 JAB85875.1 GGFJ01008016 MBW57157.1 GGFJ01008047 MBW57188.1 GFPF01008563 MAA19709.1 GADI01004007 JAA69801.1 AK417493 BAN20708.1 GDHF01026700 JAI25614.1 EAA12759.4 UFQS01000180 UFQT01000180 SSX00826.1 SSX21206.1 CH933807 EDW12893.2 GACK01004491 JAA60543.1 GGMS01017526 MBY86729.1 GACK01002275 JAA62759.1 MNPL01008544 OQR74180.1 APCN01000774 ABJB010175462 ABJB010273414 ABJB010820902 DS945857 EEC18618.1 QOIP01000005 RLU22378.1 KK119043 KFM74486.1 KK107102 EZA59607.1 GACK01008396 JAA56638.1
Proteomes
UP000276569
UP000007151
UP000053268
UP000053240
UP000007798
UP000007801
+ More
UP000192223 UP000008711 UP000235965 UP000008820 UP000000803 UP000001292 UP000075885 UP000019118 UP000030742 UP000076407 UP000009046 UP000069272 UP000007266 UP000192221 UP000002282 UP000095300 UP000001819 UP000092553 UP000007062 UP000009192 UP000192247 UP000075840 UP000001555 UP000279307 UP000054359 UP000053097 UP000075884 UP000075882
UP000192223 UP000008711 UP000235965 UP000008820 UP000000803 UP000001292 UP000075885 UP000019118 UP000030742 UP000076407 UP000009046 UP000069272 UP000007266 UP000192221 UP000002282 UP000095300 UP000001819 UP000092553 UP000007062 UP000009192 UP000192247 UP000075840 UP000001555 UP000279307 UP000054359 UP000053097 UP000075884 UP000075882
Interpro
SUPFAM
SSF48726
SSF48726
Gene 3D
ProteinModelPortal
A0A2W1BRF1
A0A3B0E3L9
A0A212F2E6
A0A0N1IN03
A0A0N1PHP0
A0A0Q9WS22
+ More
A0A1B6FKJ1 B3MMX2 A0A1B6M8H5 A0A1W4WZJ0 A0A1Y1LSW9 A0A1B6LW55 T1E289 A0A0Q5WBY4 A0A0J9QXX0 U5EQY7 T1DEN1 A0A2J7R3K5 Q16ML7 T1D4G7 Q16ML6 Q961I4 A0A0J9QZ03 A0A023ENP6 Q8IPG9 A0A0J9QYA8 A0A0J9TIG8 D3TLY5 B4HYE5 M9PCW5 Q7KTJ7 A0A2M4ABL6 A0A2M4AB32 A0A182PLT5 A0A2M4DNX6 A0A2M4DMI6 A0A0J9QXT7 N6SRJ7 A0A2M4AB34 A0A2M4AAX2 A0A2P6LA76 V5GVB3 A0A0A1XRN1 A0A182XFV9 A0A0J9QYA0 T1DHN0 A0A0J9TIH2 A0A0A9W6U6 A0A2M4DMK3 A0A1L8DA04 A0A1S4GY62 A0A1B6D4E0 E0VQ49 A0A1W7RA30 A0A182F3H9 A0A1B6DMQ0 A0A2M3Z483 A0A1Q3G3R7 A0A2M4DMQ8 A0A0A9X5C1 D6WQV4 A0A1Q3FY83 A0A1Q3FYB0 A0A2M3ZE27 A0A0N8ES29 A0A1W4VWC6 A0A0R1DT06 A0A1I8NUC4 Q29L42 A0A1Q3FY55 A0A0M4E994 W8AD12 A0A2M4BVQ8 A0A2M4BWT8 A0A224YVE7 A0A0K8RFE5 R4WD95 A0A0K8UG24 Q7Q4I1 A0A336LT56 B4KEC5 L7MBI1 A0A2S2R9Q4 L7MHV3 A0A1V9XKW6 A0A2C9GPZ7 B7QIE6 A0A3L8DRF8 A0A2C9GQ66 A0A087UAU7 A0A026WUC4 A0A182MYY3 A0A2C9GQ04 A0A182L0G9 L7M0I3
A0A1B6FKJ1 B3MMX2 A0A1B6M8H5 A0A1W4WZJ0 A0A1Y1LSW9 A0A1B6LW55 T1E289 A0A0Q5WBY4 A0A0J9QXX0 U5EQY7 T1DEN1 A0A2J7R3K5 Q16ML7 T1D4G7 Q16ML6 Q961I4 A0A0J9QZ03 A0A023ENP6 Q8IPG9 A0A0J9QYA8 A0A0J9TIG8 D3TLY5 B4HYE5 M9PCW5 Q7KTJ7 A0A2M4ABL6 A0A2M4AB32 A0A182PLT5 A0A2M4DNX6 A0A2M4DMI6 A0A0J9QXT7 N6SRJ7 A0A2M4AB34 A0A2M4AAX2 A0A2P6LA76 V5GVB3 A0A0A1XRN1 A0A182XFV9 A0A0J9QYA0 T1DHN0 A0A0J9TIH2 A0A0A9W6U6 A0A2M4DMK3 A0A1L8DA04 A0A1S4GY62 A0A1B6D4E0 E0VQ49 A0A1W7RA30 A0A182F3H9 A0A1B6DMQ0 A0A2M3Z483 A0A1Q3G3R7 A0A2M4DMQ8 A0A0A9X5C1 D6WQV4 A0A1Q3FY83 A0A1Q3FYB0 A0A2M3ZE27 A0A0N8ES29 A0A1W4VWC6 A0A0R1DT06 A0A1I8NUC4 Q29L42 A0A1Q3FY55 A0A0M4E994 W8AD12 A0A2M4BVQ8 A0A2M4BWT8 A0A224YVE7 A0A0K8RFE5 R4WD95 A0A0K8UG24 Q7Q4I1 A0A336LT56 B4KEC5 L7MBI1 A0A2S2R9Q4 L7MHV3 A0A1V9XKW6 A0A2C9GPZ7 B7QIE6 A0A3L8DRF8 A0A2C9GQ66 A0A087UAU7 A0A026WUC4 A0A182MYY3 A0A2C9GQ04 A0A182L0G9 L7M0I3
PDB
4U0Q
E-value=3.47232e-13,
Score=180
Ontologies
GO
Topology
SignalP
Position: 1 - 32,
Likelihood: 0.962186
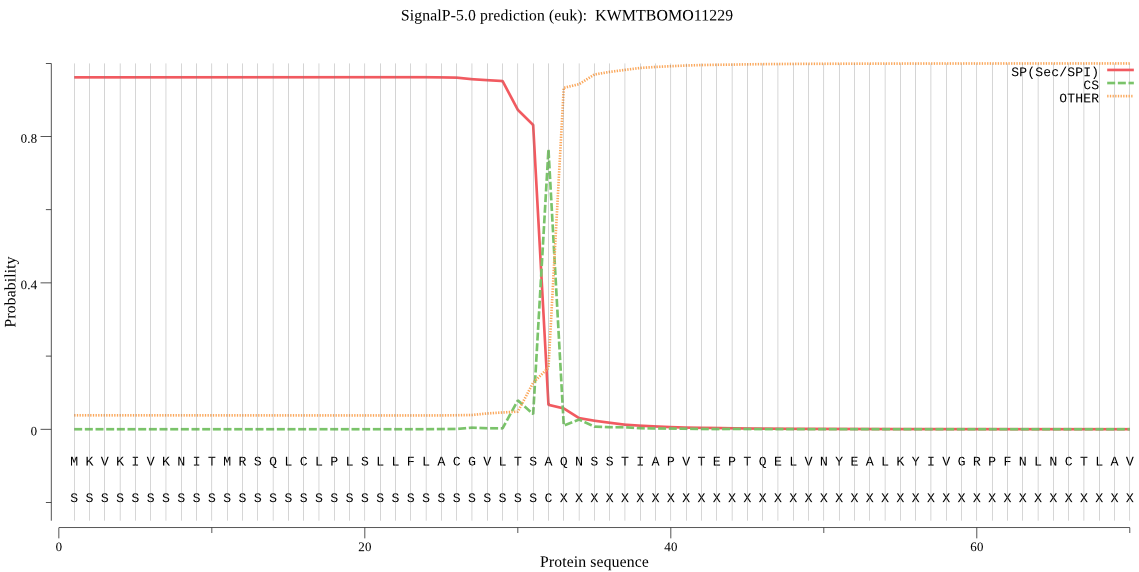
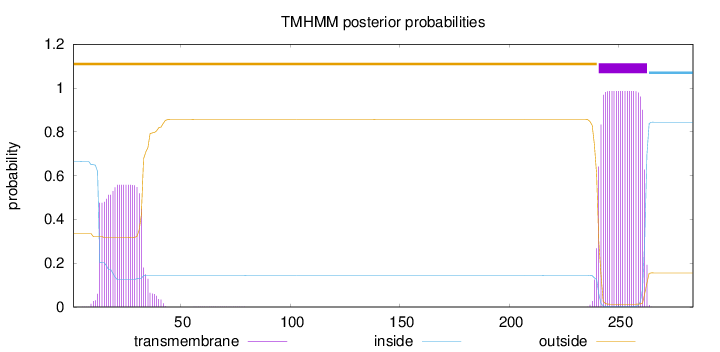
Length:
284
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
32.824
Exp number, first 60 AAs:
11.47796
Total prob of N-in:
0.66419
POSSIBLE N-term signal
sequence
outside
1 - 240
TMhelix
241 - 263
inside
264 - 284
Population Genetic Test Statistics
Pi
188.682374
Theta
182.161127
Tajima's D
-0.860792
CLR
0.87125
CSRT
0.165441727913604
Interpretation
Uncertain
