Gene
KWMTBOMO11226
Pre Gene Modal
BGIBMGA003844
Annotation
PREDICTED:_nose_resistant_to_fluoxetine_protein_6-like_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.802
Sequence
CDS
ATGTCCGAAATTCGTGACTGCAGAAAATATTGGTGGCAACATCTCTTGTACATCCACAACTACACTAAAGACTCAAACTGCTTGTTACAATCTTGGTACATAGCCGCTGATGTGCAGCTATTCATTTTTGGATTAATCATCTATCTAGTCAGCCGAAATCCAAGATCTAGGAAGATAATCCTCCCAACTGTTTTCTTCATCGGCATTGCCATTATAGCAGCCCACACCTACTTGGAAGACTTGGACGGTACCGTCATGGTTACTCCTGAAGTCATTCGTAACCAAATCAGGAGAGTCCCGACGTTCTTAAAAGTGTACAGACGAAGTCACACTAATATACCCTGTTATATCCTGGGGATGGGTGCAGGATACCTGTTTTACTATTGGCAGAAGATTAATCTGAATTTGCACAAATTTAAGAAATATAAATTGCTCTGCTGCATGCTTGGTCCGCTGCCAGTAGTCCTAGGTTCTGGCATCATGGTCACGGCCTCGTATTTCTACATGGACGCGCCAAGTTCTTATATGCGCAATGATCATGAAACTTGA
Protein
MSEIRDCRKYWWQHLLYIHNYTKDSNCLLQSWYIAADVQLFIFGLIIYLVSRNPRSRKIILPTVFFIGIAIIAAHTYLEDLDGTVMVTPEVIRNQIRRVPTFLKVYRRSHTNIPCYILGMGAGYLFYYWQKINLNLHKFKKYKLLCCMLGPLPVVLGSGIMVTASYFYMDAPSSYMRNDHET
Summary
Uniprot
H9J2V7
A0A2H1WAR4
A0A0L7LJT8
A0A2W1BKH3
A0A2A4JGE1
A0A194QFD0
+ More
A0A2A4JGW2 A0A2W1BK62 A0A2H1WAT5 A0A0L7L068 A0A0N1I4J5 A0A212EPG8 A0A0L7KZR2 A0A0N0PAP1 A0A212EGP9 A0A194QEZ9 A0A212ETZ7 A0A3S2P8D1 A0A2A4JGF7 A0A194QF19 A0A2W1BKB8 A0A2H1WAT4 A0A194QKZ1 A0A212EME8 A0A194QGQ7 A0A2A4JF89 A0A194QF64 A0A0N1IBJ6 H9J531 A0A084WEE0 A0A182IST2 A0A0N0P9S1 A0A2A4JFY4 A0A1B0DIG0 A0A1B0CI80 A0A0N1ID86 A0A336LJU7 A0A1Y9J0F2 A0A182UXI0 A0A1Y1KVX9 A0A023ESA0 A0A194Q6D5 Q7PTG8 A0A182JDV2 A0A1Y9GKR7 A0A1Y9GKR1 A0A084W2G5 A0A182I2D8 A0A0L0CN36 A0A2J7Q6G4 A0A182TJQ2 A0A0N0PCP5 A0A182QX25 A0A182SAM1 A0A182MH94 A0A1Y9GKS0 A0NBK6 A7URR5 A0A336N0M2 Q16E87 A0A182KQ81 A0A182FDU4 A0A182J629 A0A1Y9IS55 A0A182JNH6 Q16SA6 A0A182WLD6 A0A1Y9J0V5 A7URR6 W5JVJ3 A0A182LZY6 A7URR7 A7URR4 A0A182WLD8 A0A182K8B2 W5JUV2 A0A084WED7 A0A182GNM9 A0A084WED6 A0A182R4K7 A0A1Y9J0V8 W5JFI4 A0A182YNQ4 A0A1Y9GL72 A0A1W4V1V5 A0A1W4VFG7 A0A182LZ52 A0A182IAC3 A0A1Y9J0F0 A0A2H1WCF8 A0NCC8 A0A182KQ86 A0A182W4K6 B3NAL9
A0A2A4JGW2 A0A2W1BK62 A0A2H1WAT5 A0A0L7L068 A0A0N1I4J5 A0A212EPG8 A0A0L7KZR2 A0A0N0PAP1 A0A212EGP9 A0A194QEZ9 A0A212ETZ7 A0A3S2P8D1 A0A2A4JGF7 A0A194QF19 A0A2W1BKB8 A0A2H1WAT4 A0A194QKZ1 A0A212EME8 A0A194QGQ7 A0A2A4JF89 A0A194QF64 A0A0N1IBJ6 H9J531 A0A084WEE0 A0A182IST2 A0A0N0P9S1 A0A2A4JFY4 A0A1B0DIG0 A0A1B0CI80 A0A0N1ID86 A0A336LJU7 A0A1Y9J0F2 A0A182UXI0 A0A1Y1KVX9 A0A023ESA0 A0A194Q6D5 Q7PTG8 A0A182JDV2 A0A1Y9GKR7 A0A1Y9GKR1 A0A084W2G5 A0A182I2D8 A0A0L0CN36 A0A2J7Q6G4 A0A182TJQ2 A0A0N0PCP5 A0A182QX25 A0A182SAM1 A0A182MH94 A0A1Y9GKS0 A0NBK6 A7URR5 A0A336N0M2 Q16E87 A0A182KQ81 A0A182FDU4 A0A182J629 A0A1Y9IS55 A0A182JNH6 Q16SA6 A0A182WLD6 A0A1Y9J0V5 A7URR6 W5JVJ3 A0A182LZY6 A7URR7 A7URR4 A0A182WLD8 A0A182K8B2 W5JUV2 A0A084WED7 A0A182GNM9 A0A084WED6 A0A182R4K7 A0A1Y9J0V8 W5JFI4 A0A182YNQ4 A0A1Y9GL72 A0A1W4V1V5 A0A1W4VFG7 A0A182LZ52 A0A182IAC3 A0A1Y9J0F0 A0A2H1WCF8 A0NCC8 A0A182KQ86 A0A182W4K6 B3NAL9
Pubmed
EMBL
BABH01043041
ODYU01007420
SOQ50171.1
JTDY01000850
KOB75675.1
KZ150085
+ More
PZC73777.1 NWSH01001667 PCG70473.1 KQ459053 KPJ04119.1 NWSH01001633 PCG70632.1 KZ150068 PZC74095.1 SOQ50170.1 JTDY01004025 KOB68696.1 KQ459449 KPJ00879.1 AGBW02013474 OWR43366.1 KOB68697.1 KQ461184 KPJ07529.1 AGBW02015045 OWR40663.1 KPJ04118.1 AGBW02012502 OWR44975.1 RSAL01000205 RVE44373.1 PCG70472.1 KPJ04014.1 PZC74094.1 SOQ50173.1 KPJ04116.1 AGBW02013856 OWR42649.1 KPJ04120.1 PCG70631.1 KPJ04122.1 KPJ07525.1 BABH01038834 ATLV01023205 KE525341 KFB48584.1 KPJ00884.1 PCG70474.1 AJVK01034397 AJWK01013083 KPJ07535.1 UFQS01000038 UFQT01000038 SSW98078.1 SSX18464.1 GEZM01074205 GEZM01074204 JAV64641.1 GAPW01001480 JAC12118.1 KQ459582 KPI98970.1 AAAB01008807 EAA03947.5 APCN01000152 APCN01000151 ATLV01019615 KE525275 KFB44409.1 APCN01000146 JRES01000156 KNC33753.1 NEVH01017478 PNF24171.1 KQ460635 KPJ13420.1 AXCN02001261 AXCM01005440 APCN01000148 APCN01000149 EAU77672.2 EDO64707.1 UFQT01006673 SSX36124.1 CH478951 EAT32544.1 CH477678 EAT37346.1 EDO64708.1 ADMH02000306 ETN67035.1 AXCM01001276 AXCM01001277 EDO64709.1 EDO64706.1 ADMH02000427 ETN66544.1 ATLV01023204 KFB48581.1 JXUM01076912 JXUM01076913 JXUM01076914 JXUM01076915 KQ562977 KXJ74719.1 KFB48580.1 ADMH02001642 ETN61640.1 AXCM01008833 APCN01001143 SOQ50174.1 AAAB01008846 EAU77249.3 CH954177 EDV59773.2
PZC73777.1 NWSH01001667 PCG70473.1 KQ459053 KPJ04119.1 NWSH01001633 PCG70632.1 KZ150068 PZC74095.1 SOQ50170.1 JTDY01004025 KOB68696.1 KQ459449 KPJ00879.1 AGBW02013474 OWR43366.1 KOB68697.1 KQ461184 KPJ07529.1 AGBW02015045 OWR40663.1 KPJ04118.1 AGBW02012502 OWR44975.1 RSAL01000205 RVE44373.1 PCG70472.1 KPJ04014.1 PZC74094.1 SOQ50173.1 KPJ04116.1 AGBW02013856 OWR42649.1 KPJ04120.1 PCG70631.1 KPJ04122.1 KPJ07525.1 BABH01038834 ATLV01023205 KE525341 KFB48584.1 KPJ00884.1 PCG70474.1 AJVK01034397 AJWK01013083 KPJ07535.1 UFQS01000038 UFQT01000038 SSW98078.1 SSX18464.1 GEZM01074205 GEZM01074204 JAV64641.1 GAPW01001480 JAC12118.1 KQ459582 KPI98970.1 AAAB01008807 EAA03947.5 APCN01000152 APCN01000151 ATLV01019615 KE525275 KFB44409.1 APCN01000146 JRES01000156 KNC33753.1 NEVH01017478 PNF24171.1 KQ460635 KPJ13420.1 AXCN02001261 AXCM01005440 APCN01000148 APCN01000149 EAU77672.2 EDO64707.1 UFQT01006673 SSX36124.1 CH478951 EAT32544.1 CH477678 EAT37346.1 EDO64708.1 ADMH02000306 ETN67035.1 AXCM01001276 AXCM01001277 EDO64709.1 EDO64706.1 ADMH02000427 ETN66544.1 ATLV01023204 KFB48581.1 JXUM01076912 JXUM01076913 JXUM01076914 JXUM01076915 KQ562977 KXJ74719.1 KFB48580.1 ADMH02001642 ETN61640.1 AXCM01008833 APCN01001143 SOQ50174.1 AAAB01008846 EAU77249.3 CH954177 EDV59773.2
Proteomes
UP000005204
UP000037510
UP000218220
UP000053268
UP000007151
UP000053240
+ More
UP000283053 UP000030765 UP000075880 UP000092462 UP000092461 UP000076407 UP000075903 UP000007062 UP000075840 UP000037069 UP000235965 UP000075902 UP000075886 UP000075901 UP000075883 UP000008820 UP000075882 UP000069272 UP000075881 UP000075920 UP000000673 UP000069940 UP000249989 UP000075900 UP000076408 UP000192221 UP000008711
UP000283053 UP000030765 UP000075880 UP000092462 UP000092461 UP000076407 UP000075903 UP000007062 UP000075840 UP000037069 UP000235965 UP000075902 UP000075886 UP000075901 UP000075883 UP000008820 UP000075882 UP000069272 UP000075881 UP000075920 UP000000673 UP000069940 UP000249989 UP000075900 UP000076408 UP000192221 UP000008711
Pfam
Interpro
IPR002656
Acyl_transf_3
+ More
IPR004119 EcKinase-like
IPR011009 Kinase-like_dom_sf
IPR015897 CHK_kinase-like
IPR008949 Isoprenoid_synthase_dom_sf
IPR000092 Polyprenyl_synt
IPR007577 GlycoTrfase_DXD_sugar-bd_CS
IPR020801 PKS_acyl_transferase
IPR016039 Thiolase-like
IPR014030 Ketoacyl_synth_N
IPR020843 PKS_ER
IPR001227 Ac_transferase_dom_sf
IPR029058 AB_hydrolase
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR023102 Fatty_acid_synthase_dom_2
IPR014031 Ketoacyl_synth_C
IPR016035 Acyl_Trfase/lysoPLipase
IPR016036 Malonyl_transacylase_ACP-bd
IPR036291 NAD(P)-bd_dom_sf
IPR014043 Acyl_transferase
IPR001031 Thioesterase
IPR032821 KAsynt_C_assoc
IPR011032 GroES-like_sf
IPR042104 PKS_dehydratase_sf
IPR006621 Nose-resist-to-fluoxetine_N
IPR034223 IGCP-like_CPD
IPR000834 Peptidase_M14
IPR004119 EcKinase-like
IPR011009 Kinase-like_dom_sf
IPR015897 CHK_kinase-like
IPR008949 Isoprenoid_synthase_dom_sf
IPR000092 Polyprenyl_synt
IPR007577 GlycoTrfase_DXD_sugar-bd_CS
IPR020801 PKS_acyl_transferase
IPR016039 Thiolase-like
IPR014030 Ketoacyl_synth_N
IPR020843 PKS_ER
IPR001227 Ac_transferase_dom_sf
IPR029058 AB_hydrolase
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR023102 Fatty_acid_synthase_dom_2
IPR014031 Ketoacyl_synth_C
IPR016035 Acyl_Trfase/lysoPLipase
IPR016036 Malonyl_transacylase_ACP-bd
IPR036291 NAD(P)-bd_dom_sf
IPR014043 Acyl_transferase
IPR001031 Thioesterase
IPR032821 KAsynt_C_assoc
IPR011032 GroES-like_sf
IPR042104 PKS_dehydratase_sf
IPR006621 Nose-resist-to-fluoxetine_N
IPR034223 IGCP-like_CPD
IPR000834 Peptidase_M14
SUPFAM
ProteinModelPortal
H9J2V7
A0A2H1WAR4
A0A0L7LJT8
A0A2W1BKH3
A0A2A4JGE1
A0A194QFD0
+ More
A0A2A4JGW2 A0A2W1BK62 A0A2H1WAT5 A0A0L7L068 A0A0N1I4J5 A0A212EPG8 A0A0L7KZR2 A0A0N0PAP1 A0A212EGP9 A0A194QEZ9 A0A212ETZ7 A0A3S2P8D1 A0A2A4JGF7 A0A194QF19 A0A2W1BKB8 A0A2H1WAT4 A0A194QKZ1 A0A212EME8 A0A194QGQ7 A0A2A4JF89 A0A194QF64 A0A0N1IBJ6 H9J531 A0A084WEE0 A0A182IST2 A0A0N0P9S1 A0A2A4JFY4 A0A1B0DIG0 A0A1B0CI80 A0A0N1ID86 A0A336LJU7 A0A1Y9J0F2 A0A182UXI0 A0A1Y1KVX9 A0A023ESA0 A0A194Q6D5 Q7PTG8 A0A182JDV2 A0A1Y9GKR7 A0A1Y9GKR1 A0A084W2G5 A0A182I2D8 A0A0L0CN36 A0A2J7Q6G4 A0A182TJQ2 A0A0N0PCP5 A0A182QX25 A0A182SAM1 A0A182MH94 A0A1Y9GKS0 A0NBK6 A7URR5 A0A336N0M2 Q16E87 A0A182KQ81 A0A182FDU4 A0A182J629 A0A1Y9IS55 A0A182JNH6 Q16SA6 A0A182WLD6 A0A1Y9J0V5 A7URR6 W5JVJ3 A0A182LZY6 A7URR7 A7URR4 A0A182WLD8 A0A182K8B2 W5JUV2 A0A084WED7 A0A182GNM9 A0A084WED6 A0A182R4K7 A0A1Y9J0V8 W5JFI4 A0A182YNQ4 A0A1Y9GL72 A0A1W4V1V5 A0A1W4VFG7 A0A182LZ52 A0A182IAC3 A0A1Y9J0F0 A0A2H1WCF8 A0NCC8 A0A182KQ86 A0A182W4K6 B3NAL9
A0A2A4JGW2 A0A2W1BK62 A0A2H1WAT5 A0A0L7L068 A0A0N1I4J5 A0A212EPG8 A0A0L7KZR2 A0A0N0PAP1 A0A212EGP9 A0A194QEZ9 A0A212ETZ7 A0A3S2P8D1 A0A2A4JGF7 A0A194QF19 A0A2W1BKB8 A0A2H1WAT4 A0A194QKZ1 A0A212EME8 A0A194QGQ7 A0A2A4JF89 A0A194QF64 A0A0N1IBJ6 H9J531 A0A084WEE0 A0A182IST2 A0A0N0P9S1 A0A2A4JFY4 A0A1B0DIG0 A0A1B0CI80 A0A0N1ID86 A0A336LJU7 A0A1Y9J0F2 A0A182UXI0 A0A1Y1KVX9 A0A023ESA0 A0A194Q6D5 Q7PTG8 A0A182JDV2 A0A1Y9GKR7 A0A1Y9GKR1 A0A084W2G5 A0A182I2D8 A0A0L0CN36 A0A2J7Q6G4 A0A182TJQ2 A0A0N0PCP5 A0A182QX25 A0A182SAM1 A0A182MH94 A0A1Y9GKS0 A0NBK6 A7URR5 A0A336N0M2 Q16E87 A0A182KQ81 A0A182FDU4 A0A182J629 A0A1Y9IS55 A0A182JNH6 Q16SA6 A0A182WLD6 A0A1Y9J0V5 A7URR6 W5JVJ3 A0A182LZY6 A7URR7 A7URR4 A0A182WLD8 A0A182K8B2 W5JUV2 A0A084WED7 A0A182GNM9 A0A084WED6 A0A182R4K7 A0A1Y9J0V8 W5JFI4 A0A182YNQ4 A0A1Y9GL72 A0A1W4V1V5 A0A1W4VFG7 A0A182LZ52 A0A182IAC3 A0A1Y9J0F0 A0A2H1WCF8 A0NCC8 A0A182KQ86 A0A182W4K6 B3NAL9
Ontologies
GO
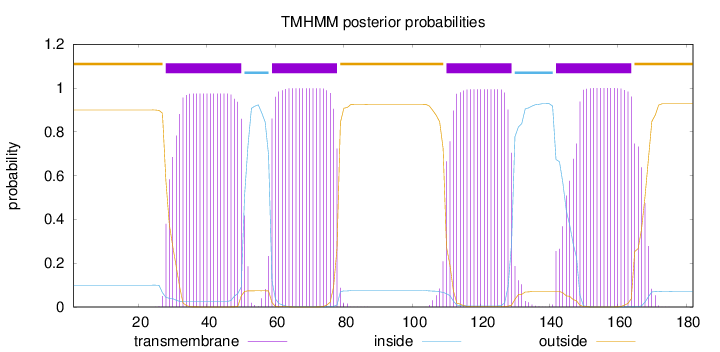
Topology
Length:
182
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
83.10518
Exp number, first 60 AAs:
23.50698
Total prob of N-in:
0.09982
POSSIBLE N-term signal
sequence
outside
1 - 27
TMhelix
28 - 50
inside
51 - 58
TMhelix
59 - 78
outside
79 - 109
TMhelix
110 - 129
inside
130 - 141
TMhelix
142 - 164
outside
165 - 182
Population Genetic Test Statistics
Pi
146.198143
Theta
138.546088
Tajima's D
-0.644449
CLR
117.833189
CSRT
0.204139793010349
Interpretation
Uncertain
