Gene
KWMTBOMO11207 Validated by peptides from experiments
Annotation
PREDICTED:_glucose_dehydrogenase_[FAD?_quinone]-like?_partial_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.951
Sequence
CDS
ATGAATACAACATCCACATTGAATGAAATTCATAGACTTCAAACCGGCTTTCAAATACTGGCAGCTCTACAGCTAACTGGGTATCTTTGGCCACAAGAAGCGGTTATTCAAGATGGTGCCAAGTATGATTTCATTGTTGTCGGCGCTGGATCTGGTGGATGCGTGGTCGCTAATAGACTGTCGGAGATTAAAAACATCAATGTATTGCTGATTGAAGCTGGAGGAGATCCCCCAATAGAATCTGTTATTCCAAGCTTAAACCTATTTATGAAGGTTTCAAAAGTAGACTGGAATTACACATCCGTCCCGGATGTAATGATGAAATACATGAAAAATCAAGTCGTCCAGGTTACAAGAGGGAAAATGCTCGGTGGGACTAGTTCTATCAATTTCATGGCCTACACGAGAGGGAATTCCCAAGATTATGATACATGGGCCAAAATTATTAATGATACAACTTGGAAGTGGGAGAACGTGCTACCGTACTTCATCAAGAGTGAAAGACTTGATGAATCCGATCTAAATTGTGTTAACAAAGAACTTCATGGTACTGAAGGTTATTTGGGCGTCAGCACACAACACAGTGATGCGACATCTGAATACTTGAAAGCTTTTTCTGAAGTCGGTCATGATATCGAATACGGGATAAATGGAGACAAACCAGTTGGTTTTAGTGAGCCATTATTTACAATTGCAAATGGAATACGACAAAGTGCTGCTTTAGCATTTCTCTCAAAATTTCAGGATCGTGCTAATCTACATGTTTTGAAGAATGCTTTAGTTACAAAAATTTTGTTCGATGACAACAAATGTGCAACTGGTGTTGAATTAATATTAAATGATAGATTGATGACAATAAGGGCAACTAAAGAAGTAATTGTGTCTGCAGGTGCTATCAACTCTCCTCAATTGTTGATGCTTTCAGGAGTTGGGCCTCGGCAGCATTTAAAACATAAAAACATCAAGGTTACATCTGATTTACCAGTAGGACAAAAAATGCAAGATCACATTAACGTTCTAGTTTTACATAAAATGGGTAAAGCTACTTTAAATAAGAAACCAGCTGATCCTTTGAAATCTCCATTACCTTTAATCTTAGGATACGCAGCTTTGAACAAGACTCAAAAGTATCCTGACTACGAGATAATGACGCTTATTATAAATGATAAGAATGATTTTATGCCTTTTTGTGTGTTTTTCTTGAGCTTAAAAGACGAAATCTGCCAAGATTATTATGACCATAGTGGTGACCGGCAATTGATGTATACATTGATGAGTTACTTATATCCGAAAGCTCGCGGCCAAATTCTGTTGAACACAACAAATGCTCGCGATCAACCTCTCATTTACCCTAACTATCTCACAGAAGAAAAAGATATAGAAAACCTTGAAAAATATGTTAAAGATTACCTCAAAGTTACGAACTCTAGCTTCTTCAAAAGTGTTAATTCAGAGCTTATAATTCCAAGGGCTTGTAACTGCGTTCCTGATCAAAATTTTTGGAAATGTTACGTGAAATGCCTGGTTATCCCTGGCTATCATTTTTCCGGTACTTGTGCTATGGGTGAAGTGGTAGATTCTAGGTTAAGAGTCTACGGAGTTAAAAAACTGAGAGTCGTGGATGCCAGTGTGATGCCCCGAATACCTGGAGCCCATACCAATGCACCAACTATAATGATCGGTGAAAAAGCAGCCGATATGATAAAAGACGAACATGAGCTGCTGTTGCTGCAAAATTTAAAACCGAATTAA
Protein
MNTTSTLNEIHRLQTGFQILAALQLTGYLWPQEAVIQDGAKYDFIVVGAGSGGCVVANRLSEIKNINVLLIEAGGDPPIESVIPSLNLFMKVSKVDWNYTSVPDVMMKYMKNQVVQVTRGKMLGGTSSINFMAYTRGNSQDYDTWAKIINDTTWKWENVLPYFIKSERLDESDLNCVNKELHGTEGYLGVSTQHSDATSEYLKAFSEVGHDIEYGINGDKPVGFSEPLFTIANGIRQSAALAFLSKFQDRANLHVLKNALVTKILFDDNKCATGVELILNDRLMTIRATKEVIVSAGAINSPQLLMLSGVGPRQHLKHKNIKVTSDLPVGQKMQDHINVLVLHKMGKATLNKKPADPLKSPLPLILGYAALNKTQKYPDYEIMTLIINDKNDFMPFCVFFLSLKDEICQDYYDHSGDRQLMYTLMSYLYPKARGQILLNTTNARDQPLIYPNYLTEEKDIENLEKYVKDYLKVTNSSFFKSVNSELIIPRACNCVPDQNFWKCYVKCLVIPGYHFSGTCAMGEVVDSRLRVYGVKKLRVVDASVMPRIPGAHTNAPTIMIGEKAADMIKDEHELLLLQNLKPN
Summary
Cofactor
FAD
Similarity
Belongs to the GMC oxidoreductase family.
Uniprot
A0A2W1BQ09
A0A0L7LL99
A0A3S2N607
A0A3S2LRK7
A0A2H1VCU6
A0A2A4JWF9
+ More
A0A2W1BRA5 A0A2H1VCV1 A0A2H1VB55 A0A212F6I8 A0A2H1VP10 D0ABA6 A0A0L7LPB4 A0A2A4K5W1 A0A2H1WHG3 A0A2A4JYC1 A0A0N0PDR9 A0A2H1WZ72 A0A2H1W336 A0A0L7LUK3 H9J868 A0A2W1BM44 A0A0N1PIV1 A0A2W1BCG7 A0A2A4JSK9 A0A194QAR0 A0A2A4K6C3 A0A2W1B6G7 A0A2A4K9G4 H9JLP6 H9J869 A0A0N1IPT1 A0A2H1V0S8 A0A2H1VDA1 A0A2W1BIV7 A0A2A4JUI0 A0A0N1I9P3 A0A2A4IZE5 A0A212F6I0 A0A194QGE4 A0A2W1BIM1 H9JLQ9 A0A194QAM1 A0A2H1WHH0 A0A0L7LQD8 A0A2H1W3T3 A0A2H1WPH8 A0A212FMS7 A0A2H1VYA6 A0A2A4IZ69 A0A2A4JQV7 A0A0L7KN60 A0A2A4K7J0 A0A0L7L247 A0A2W1BPR7 A0A2W1BHP2 A0A194PPA0 A0A2H1VZZ7 C7EPE2 A0A2A4JF29 A0A0L7KY98 H9IS42 A0A2W1BU23 A0A2H1WQ34 A0A2W1BFD2 A0A194QNE6 A0A2W1BEK9 A0A2A4JE08 A0A2H1W7M4 A0A0L7L6H5 H9JLW3 A0A088S376 A0A088SM07 A0A088S3T2 A0A088S3M8 A0A088S5A0 A0A2H1W0I8 A0A088S3S6 A0A0L7KKV3 H6AGY0 A0A088SM03 B8Y866 A0A212EII3 A0A088S3M1 A0A2A4J139 A0A0N0PE61 A0A194R898 A0A194QC04 A0A182FZT3 A0A0L7LP50 A0A194PXZ3 A0A182FZT5 A0A1B6HJ69 A0A1B6LHN9 A0A1B0D7C5 A0A2H1V9N6 A0A1Q3FZM7 A0A1B6EYD4 A0A2W1B5P9 A0A154PBS2
A0A2W1BRA5 A0A2H1VCV1 A0A2H1VB55 A0A212F6I8 A0A2H1VP10 D0ABA6 A0A0L7LPB4 A0A2A4K5W1 A0A2H1WHG3 A0A2A4JYC1 A0A0N0PDR9 A0A2H1WZ72 A0A2H1W336 A0A0L7LUK3 H9J868 A0A2W1BM44 A0A0N1PIV1 A0A2W1BCG7 A0A2A4JSK9 A0A194QAR0 A0A2A4K6C3 A0A2W1B6G7 A0A2A4K9G4 H9JLP6 H9J869 A0A0N1IPT1 A0A2H1V0S8 A0A2H1VDA1 A0A2W1BIV7 A0A2A4JUI0 A0A0N1I9P3 A0A2A4IZE5 A0A212F6I0 A0A194QGE4 A0A2W1BIM1 H9JLQ9 A0A194QAM1 A0A2H1WHH0 A0A0L7LQD8 A0A2H1W3T3 A0A2H1WPH8 A0A212FMS7 A0A2H1VYA6 A0A2A4IZ69 A0A2A4JQV7 A0A0L7KN60 A0A2A4K7J0 A0A0L7L247 A0A2W1BPR7 A0A2W1BHP2 A0A194PPA0 A0A2H1VZZ7 C7EPE2 A0A2A4JF29 A0A0L7KY98 H9IS42 A0A2W1BU23 A0A2H1WQ34 A0A2W1BFD2 A0A194QNE6 A0A2W1BEK9 A0A2A4JE08 A0A2H1W7M4 A0A0L7L6H5 H9JLW3 A0A088S376 A0A088SM07 A0A088S3T2 A0A088S3M8 A0A088S5A0 A0A2H1W0I8 A0A088S3S6 A0A0L7KKV3 H6AGY0 A0A088SM03 B8Y866 A0A212EII3 A0A088S3M1 A0A2A4J139 A0A0N0PE61 A0A194R898 A0A194QC04 A0A182FZT3 A0A0L7LP50 A0A194PXZ3 A0A182FZT5 A0A1B6HJ69 A0A1B6LHN9 A0A1B0D7C5 A0A2H1V9N6 A0A1Q3FZM7 A0A1B6EYD4 A0A2W1B5P9 A0A154PBS2
EMBL
KZ150085
PZC73783.1
JTDY01000758
KOB75991.1
RSAL01000341
RVE42398.1
+ More
RVE42399.1 ODYU01001603 SOQ38054.1 NWSH01000445 PCG76355.1 KZ149986 PZC75677.1 ODYU01001607 SOQ38064.1 SOQ38065.1 AGBW02010007 OWR49347.1 ODYU01003587 SOQ42538.1 FP102341 CBH09301.1 JTDY01000418 KOB77282.1 NWSH01000095 PCG79621.1 ODYU01008704 SOQ52509.1 NWSH01000427 PCG76483.1 KQ460044 KPJ18288.1 ODYU01012146 SOQ58297.1 ODYU01006017 SOQ47491.1 JTDY01000066 KOB79079.1 BABH01039855 BABH01039856 PZC73780.1 KPJ18290.1 KZ150303 PZC71524.1 NWSH01000760 PCG74382.1 KQ459232 KPJ02519.1 PCG79619.1 KZ150274 PZC71662.1 NWSH01000039 PCG80370.1 BABH01005285 BABH01039860 BABH01039861 KPJ18289.1 ODYU01000155 SOQ34448.1 ODYU01001935 SOQ38829.1 KZ150015 PZC74989.1 NWSH01000559 PCG75661.1 KPJ18287.1 NWSH01004349 PCG65195.1 OWR49345.1 KPJ02521.1 PZC71523.1 BABH01005224 BABH01005225 BABH01005226 KPJ02522.1 SOQ52508.1 JTDY01000335 KOB77647.1 SOQ47492.1 ODYU01010073 SOQ54917.1 AGBW02007651 OWR55045.1 ODYU01005196 SOQ45815.1 NWSH01004322 PCG65227.1 PCG74381.1 JTDY01008771 KOB64409.1 PCG79620.1 JTDY01003559 KOB69344.1 KZ149963 PZC76281.1 PZC73781.1 KQ459603 KPI92960.1 ODYU01005459 SOQ46343.1 BABH01024320 GQ285072 ACT66690.2 NWSH01001625 PCG70657.1 JTDY01004415 KOB68187.1 BABH01024325 PZC76280.1 ODYU01010216 SOQ55185.1 PZC73782.1 KQ461194 KPJ06879.1 KZ150336 PZC71326.1 NWSH01001885 PCG69794.1 ODYU01006844 SOQ49058.1 JTDY01002689 KOB70914.1 BABH01005279 KF717658 AIO02860.1 KF717656 KF717657 KF717660 KF717667 AIO02858.1 AIO02859.1 AIO02862.1 AIO02869.1 KF717661 AIO02863.1 KF717664 AIO02866.1 KF717655 AIO02857.1 SOQ46342.1 KF717651 AIO02853.1 JTDY01009199 KOB63957.1 JF433972 KF717653 KF717659 KF717662 KF717663 KF717665 KF717666 KF717668 AEM17059.1 AIO02855.1 AIO02861.1 AIO02864.1 AIO02865.1 AIO02867.1 AIO02868.1 AIO02870.1 KF717652 AIO02854.1 FJ493468 ACL36977.1 AGBW02014643 OWR41285.1 KF717654 AIO02856.1 NWSH01004424 PCG65132.1 KPJ18306.1 KQ460627 KPJ13480.1 KPJ02520.1 JTDY01000479 KOB76996.1 KQ459585 KPI98211.1 GECU01033045 JAS74661.1 GEBQ01016745 JAT23232.1 AJVK01026844 AJVK01026845 AJVK01026846 ODYU01001405 SOQ37539.1 GFDL01002031 JAV33014.1 GECZ01026787 JAS42982.1 KZ150306 PZC71499.1 KQ434869 KZC09325.1
RVE42399.1 ODYU01001603 SOQ38054.1 NWSH01000445 PCG76355.1 KZ149986 PZC75677.1 ODYU01001607 SOQ38064.1 SOQ38065.1 AGBW02010007 OWR49347.1 ODYU01003587 SOQ42538.1 FP102341 CBH09301.1 JTDY01000418 KOB77282.1 NWSH01000095 PCG79621.1 ODYU01008704 SOQ52509.1 NWSH01000427 PCG76483.1 KQ460044 KPJ18288.1 ODYU01012146 SOQ58297.1 ODYU01006017 SOQ47491.1 JTDY01000066 KOB79079.1 BABH01039855 BABH01039856 PZC73780.1 KPJ18290.1 KZ150303 PZC71524.1 NWSH01000760 PCG74382.1 KQ459232 KPJ02519.1 PCG79619.1 KZ150274 PZC71662.1 NWSH01000039 PCG80370.1 BABH01005285 BABH01039860 BABH01039861 KPJ18289.1 ODYU01000155 SOQ34448.1 ODYU01001935 SOQ38829.1 KZ150015 PZC74989.1 NWSH01000559 PCG75661.1 KPJ18287.1 NWSH01004349 PCG65195.1 OWR49345.1 KPJ02521.1 PZC71523.1 BABH01005224 BABH01005225 BABH01005226 KPJ02522.1 SOQ52508.1 JTDY01000335 KOB77647.1 SOQ47492.1 ODYU01010073 SOQ54917.1 AGBW02007651 OWR55045.1 ODYU01005196 SOQ45815.1 NWSH01004322 PCG65227.1 PCG74381.1 JTDY01008771 KOB64409.1 PCG79620.1 JTDY01003559 KOB69344.1 KZ149963 PZC76281.1 PZC73781.1 KQ459603 KPI92960.1 ODYU01005459 SOQ46343.1 BABH01024320 GQ285072 ACT66690.2 NWSH01001625 PCG70657.1 JTDY01004415 KOB68187.1 BABH01024325 PZC76280.1 ODYU01010216 SOQ55185.1 PZC73782.1 KQ461194 KPJ06879.1 KZ150336 PZC71326.1 NWSH01001885 PCG69794.1 ODYU01006844 SOQ49058.1 JTDY01002689 KOB70914.1 BABH01005279 KF717658 AIO02860.1 KF717656 KF717657 KF717660 KF717667 AIO02858.1 AIO02859.1 AIO02862.1 AIO02869.1 KF717661 AIO02863.1 KF717664 AIO02866.1 KF717655 AIO02857.1 SOQ46342.1 KF717651 AIO02853.1 JTDY01009199 KOB63957.1 JF433972 KF717653 KF717659 KF717662 KF717663 KF717665 KF717666 KF717668 AEM17059.1 AIO02855.1 AIO02861.1 AIO02864.1 AIO02865.1 AIO02867.1 AIO02868.1 AIO02870.1 KF717652 AIO02854.1 FJ493468 ACL36977.1 AGBW02014643 OWR41285.1 KF717654 AIO02856.1 NWSH01004424 PCG65132.1 KPJ18306.1 KQ460627 KPJ13480.1 KPJ02520.1 JTDY01000479 KOB76996.1 KQ459585 KPI98211.1 GECU01033045 JAS74661.1 GEBQ01016745 JAT23232.1 AJVK01026844 AJVK01026845 AJVK01026846 ODYU01001405 SOQ37539.1 GFDL01002031 JAV33014.1 GECZ01026787 JAS42982.1 KZ150306 PZC71499.1 KQ434869 KZC09325.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A2W1BQ09
A0A0L7LL99
A0A3S2N607
A0A3S2LRK7
A0A2H1VCU6
A0A2A4JWF9
+ More
A0A2W1BRA5 A0A2H1VCV1 A0A2H1VB55 A0A212F6I8 A0A2H1VP10 D0ABA6 A0A0L7LPB4 A0A2A4K5W1 A0A2H1WHG3 A0A2A4JYC1 A0A0N0PDR9 A0A2H1WZ72 A0A2H1W336 A0A0L7LUK3 H9J868 A0A2W1BM44 A0A0N1PIV1 A0A2W1BCG7 A0A2A4JSK9 A0A194QAR0 A0A2A4K6C3 A0A2W1B6G7 A0A2A4K9G4 H9JLP6 H9J869 A0A0N1IPT1 A0A2H1V0S8 A0A2H1VDA1 A0A2W1BIV7 A0A2A4JUI0 A0A0N1I9P3 A0A2A4IZE5 A0A212F6I0 A0A194QGE4 A0A2W1BIM1 H9JLQ9 A0A194QAM1 A0A2H1WHH0 A0A0L7LQD8 A0A2H1W3T3 A0A2H1WPH8 A0A212FMS7 A0A2H1VYA6 A0A2A4IZ69 A0A2A4JQV7 A0A0L7KN60 A0A2A4K7J0 A0A0L7L247 A0A2W1BPR7 A0A2W1BHP2 A0A194PPA0 A0A2H1VZZ7 C7EPE2 A0A2A4JF29 A0A0L7KY98 H9IS42 A0A2W1BU23 A0A2H1WQ34 A0A2W1BFD2 A0A194QNE6 A0A2W1BEK9 A0A2A4JE08 A0A2H1W7M4 A0A0L7L6H5 H9JLW3 A0A088S376 A0A088SM07 A0A088S3T2 A0A088S3M8 A0A088S5A0 A0A2H1W0I8 A0A088S3S6 A0A0L7KKV3 H6AGY0 A0A088SM03 B8Y866 A0A212EII3 A0A088S3M1 A0A2A4J139 A0A0N0PE61 A0A194R898 A0A194QC04 A0A182FZT3 A0A0L7LP50 A0A194PXZ3 A0A182FZT5 A0A1B6HJ69 A0A1B6LHN9 A0A1B0D7C5 A0A2H1V9N6 A0A1Q3FZM7 A0A1B6EYD4 A0A2W1B5P9 A0A154PBS2
A0A2W1BRA5 A0A2H1VCV1 A0A2H1VB55 A0A212F6I8 A0A2H1VP10 D0ABA6 A0A0L7LPB4 A0A2A4K5W1 A0A2H1WHG3 A0A2A4JYC1 A0A0N0PDR9 A0A2H1WZ72 A0A2H1W336 A0A0L7LUK3 H9J868 A0A2W1BM44 A0A0N1PIV1 A0A2W1BCG7 A0A2A4JSK9 A0A194QAR0 A0A2A4K6C3 A0A2W1B6G7 A0A2A4K9G4 H9JLP6 H9J869 A0A0N1IPT1 A0A2H1V0S8 A0A2H1VDA1 A0A2W1BIV7 A0A2A4JUI0 A0A0N1I9P3 A0A2A4IZE5 A0A212F6I0 A0A194QGE4 A0A2W1BIM1 H9JLQ9 A0A194QAM1 A0A2H1WHH0 A0A0L7LQD8 A0A2H1W3T3 A0A2H1WPH8 A0A212FMS7 A0A2H1VYA6 A0A2A4IZ69 A0A2A4JQV7 A0A0L7KN60 A0A2A4K7J0 A0A0L7L247 A0A2W1BPR7 A0A2W1BHP2 A0A194PPA0 A0A2H1VZZ7 C7EPE2 A0A2A4JF29 A0A0L7KY98 H9IS42 A0A2W1BU23 A0A2H1WQ34 A0A2W1BFD2 A0A194QNE6 A0A2W1BEK9 A0A2A4JE08 A0A2H1W7M4 A0A0L7L6H5 H9JLW3 A0A088S376 A0A088SM07 A0A088S3T2 A0A088S3M8 A0A088S5A0 A0A2H1W0I8 A0A088S3S6 A0A0L7KKV3 H6AGY0 A0A088SM03 B8Y866 A0A212EII3 A0A088S3M1 A0A2A4J139 A0A0N0PE61 A0A194R898 A0A194QC04 A0A182FZT3 A0A0L7LP50 A0A194PXZ3 A0A182FZT5 A0A1B6HJ69 A0A1B6LHN9 A0A1B0D7C5 A0A2H1V9N6 A0A1Q3FZM7 A0A1B6EYD4 A0A2W1B5P9 A0A154PBS2
PDB
4YNU
E-value=3.02847e-48,
Score=486
Ontologies
GO
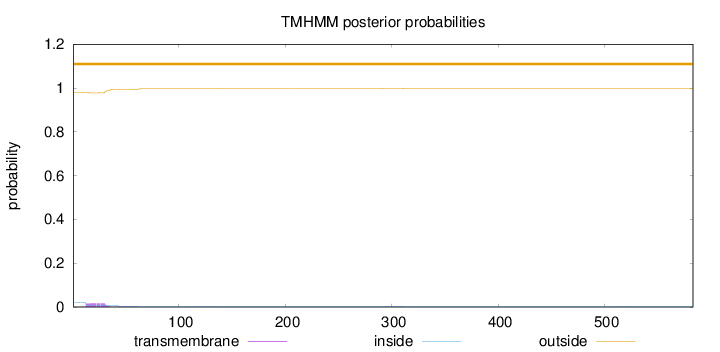
Topology
Length:
583
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.432700000000001
Exp number, first 60 AAs:
0.38516
Total prob of N-in:
0.02116
outside
1 - 583
Population Genetic Test Statistics
Pi
179.326457
Theta
192.780943
Tajima's D
1.24216
CLR
0
CSRT
0.728363581820909
Interpretation
Uncertain
