Gene
KWMTBOMO11202 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012665
Annotation
PREDICTED:_F-actin-capping_protein_subunit_alpha_[Papilio_machaon]
Full name
F-actin-capping protein subunit alpha
Location in the cell
Cytoplasmic Reliability : 2.393
Sequence
CDS
ATGGCGGCGGACGGTGACGAAGTGATTAGTGATCAAGAAAAAGTTAGAATCGTTTCAGATTTCATTTTGCATTCGCCGCCAGGGGAATTTAACGAAGTTTTCAATGATGTGAGGGTGTTGCTGAATAACGATACCTTGCTCAAGGAAGGTGCTAGTGGAGCCTTCGCTCAATACAACAAGGATCAACTCACGCCCGTACGTCTCGAAGGCTCCGAGCTGTATACACTCATCACCGACCACAACGAACTTGGTGGCGGAAGGTTTTTCGATCCTCGCTCCAAGTGTTCTTTCAGGTATGACCATCTACGCAAAGAAGCGTCCGAGTATGAACCCTACGAGCCCGATCGAGCGGCTGAGCCTTGGCGGGCGGCACTTGATGAGGAGCTAACGGCGTACGTGGCTGCACACTACAAGCATGGTGCTAGTTTAGTGGTGGGTCGCACAGTCGACTCTGGTACGGTACAGCTGGTCGCCTGTATCGAAGATCACCAGTTTCAACCCAAGAACTATTGGAACGGTCGGTGGCGTTCGGTGTGGTCTTTGACGGTCGGCGGGCCTGCTACAGAGTTACGTGGAACTCTACGTGTTCAAGTTCATTATTATGAGGATGGTAATGTACAACTTGTGAGCTCAAAGGAAATTAAGGCTCCTCTGGTGGCTAGCGGTGAAGTCGCCACTGCTAAAGAGTTCGTGCGGCTTGTCTCTGATGCAGAAAATACGTATCAAACTGCCATTTCAGACAATTACAAGACCATGAGCGACACAACGTTCAAAGCACTGAGACGACAACTACCTGTGACCCGCAGCAAGATTGACTGGGCTCGTCTTGTTTCTTACACGATCGGTAAGGAGCTCAAGAGCCAATGA
Protein
MAADGDEVISDQEKVRIVSDFILHSPPGEFNEVFNDVRVLLNNDTLLKEGASGAFAQYNKDQLTPVRLEGSELYTLITDHNELGGGRFFDPRSKCSFRYDHLRKEASEYEPYEPDRAAEPWRAALDEELTAYVAAHYKHGASLVVGRTVDSGTVQLVACIEDHQFQPKNYWNGRWRSVWSLTVGGPATELRGTLRVQVHYYEDGNVQLVSSKEIKAPLVASGEVATAKEFVRLVSDAENTYQTAISDNYKTMSDTTFKALRRQLPVTRSKIDWARLVSYTIGKELKSQ
Summary
Description
F-actin-capping proteins bind in a Ca(2+)-independent manner to the fast growing ends of actin filaments (barbed end) thereby blocking the exchange of subunits at these ends. Unlike other capping proteins (such as gelsolin and severin), these proteins do not sever actin filaments.
F-actin-capping proteins bind in a Ca(2+)-independent manner to the fast growing ends of actin filaments (barbed end) thereby blocking the exchange of subunits at these ends. Unlike other capping proteins (such as gelsolin and severin), these proteins do not sever actin filaments (By similarity).
F-actin-capping proteins bind in a Ca(2+)-independent manner to the fast growing ends of actin filaments (barbed end) thereby blocking the exchange of subunits at these ends. Unlike other capping proteins (such as gelsolin and severin), these proteins do not sever actin filaments (By similarity).
Subunit
Heterodimer of an alpha and a beta subunit.
Similarity
Belongs to the F-actin-capping protein alpha subunit family.
Keywords
Actin capping
Actin-binding
Complete proteome
Reference proteome
Feature
chain F-actin-capping protein subunit alpha
Uniprot
H9JT03
A0A194QWZ9
A0A194QF17
A0A1E1WDM6
A0A2H1WS37
A0A2W1BQV7
+ More
A0A2A4J7F6 A0A0L7LJ82 A0A212FGF2 S4PAE9 D6WRI1 A0A088A627 A0A0L7R1Z8 A0A154P3S0 E2B5B7 A0A151HZP1 A0A158NC62 E9IM12 A0A195F9A8 A0A195CN90 A0A151XCQ4 A0A1B6HQG9 A0A1B6LHE9 A0A310SN37 A0A0J7NIW0 A0A1W4XP49 E2AYP3 A0A026WVL7 V5GVA1 A0A0C9Q5X5 J3JVU5 N6UF55 A0A1Y1K5N3 A0A232EJF2 A0A067QYV3 A0A0K8TNW5 K7IR84 A0A2J7PX86 A0A0L0C5M5 T1PHZ2 A0A1I8P8L5 U5EU77 Q16I64 A0A023EMK3 B4I7I4 B4QFL0 Q9W2N0 B3NK27 B4P9J2 A0A1W4V5X5 B3MDV9 A0A0P5PYC3 E9HL07 A0A0M3QUE6 A0A1L8E5K2 B4NN68 A0A1A9V626 F4WYV2 A0A1Q3EUF1 A0A1B0ARK0 A0A182PDR9 A0A1A9WHV4 A0A182VAZ1 A0A182KNZ2 Q7Q5Y1 A0A182RQX4 A0A1J1HI91 A0A084WER0 A0A0P4VN78 A0A182Y2R4 A0A182WJX1 A0A0P6I4S5 A0A182IMQ9 E0W2S1 W8BRT2 B4LNG9 A0A0T6B9C2 A0A0K8UH13 A0A182MM38 A0A034WVC3 A0A182QSV4 A0A0A1WRX5 A0A2I9LP04 A0A3R7QCH9 T1E2U7 B4H4Y4 Q28X29 T1JNV6 A0A3B0J161 T1DFJ8 A0A1D2NGM3 A0A182K4R5 A0A2M4AF83 A0A0L8GNQ0 A0A131XVK7 A0A2L2XWT3 A0A1S3JR53 A0A1S3D469 A0A182FEI7 A0A2M4CSF0
A0A2A4J7F6 A0A0L7LJ82 A0A212FGF2 S4PAE9 D6WRI1 A0A088A627 A0A0L7R1Z8 A0A154P3S0 E2B5B7 A0A151HZP1 A0A158NC62 E9IM12 A0A195F9A8 A0A195CN90 A0A151XCQ4 A0A1B6HQG9 A0A1B6LHE9 A0A310SN37 A0A0J7NIW0 A0A1W4XP49 E2AYP3 A0A026WVL7 V5GVA1 A0A0C9Q5X5 J3JVU5 N6UF55 A0A1Y1K5N3 A0A232EJF2 A0A067QYV3 A0A0K8TNW5 K7IR84 A0A2J7PX86 A0A0L0C5M5 T1PHZ2 A0A1I8P8L5 U5EU77 Q16I64 A0A023EMK3 B4I7I4 B4QFL0 Q9W2N0 B3NK27 B4P9J2 A0A1W4V5X5 B3MDV9 A0A0P5PYC3 E9HL07 A0A0M3QUE6 A0A1L8E5K2 B4NN68 A0A1A9V626 F4WYV2 A0A1Q3EUF1 A0A1B0ARK0 A0A182PDR9 A0A1A9WHV4 A0A182VAZ1 A0A182KNZ2 Q7Q5Y1 A0A182RQX4 A0A1J1HI91 A0A084WER0 A0A0P4VN78 A0A182Y2R4 A0A182WJX1 A0A0P6I4S5 A0A182IMQ9 E0W2S1 W8BRT2 B4LNG9 A0A0T6B9C2 A0A0K8UH13 A0A182MM38 A0A034WVC3 A0A182QSV4 A0A0A1WRX5 A0A2I9LP04 A0A3R7QCH9 T1E2U7 B4H4Y4 Q28X29 T1JNV6 A0A3B0J161 T1DFJ8 A0A1D2NGM3 A0A182K4R5 A0A2M4AF83 A0A0L8GNQ0 A0A131XVK7 A0A2L2XWT3 A0A1S3JR53 A0A1S3D469 A0A182FEI7 A0A2M4CSF0
Pubmed
19121390
26354079
28756777
26227816
22118469
23622113
+ More
18362917 19820115 20798317 21347285 21282665 24508170 30249741 22516182 23537049 28004739 28648823 24845553 26369729 20075255 26108605 25315136 17510324 24945155 17994087 22936249 10731132 12537572 12537569 17550304 21292972 21719571 20966253 12364791 14747013 17210077 24438588 25244985 20566863 24495485 25348373 25830018 29248469 24330624 15632085 27289101 26561354
18362917 19820115 20798317 21347285 21282665 24508170 30249741 22516182 23537049 28004739 28648823 24845553 26369729 20075255 26108605 25315136 17510324 24945155 17994087 22936249 10731132 12537572 12537569 17550304 21292972 21719571 20966253 12364791 14747013 17210077 24438588 25244985 20566863 24495485 25348373 25830018 29248469 24330624 15632085 27289101 26561354
EMBL
BABH01038957
KQ461073
KPJ09485.1
KQ459053
KPJ04138.1
GDQN01005971
+ More
JAT85083.1 ODYU01010630 SOQ55871.1 KZ150068 PZC74113.1 NWSH01002895 PCG67352.1 JTDY01000905 KOB75490.1 AGBW02008682 OWR52807.1 GAIX01003294 JAA89266.1 KQ971351 EFA06446.1 KQ414667 KOC64843.1 KQ434809 KZC06589.1 GL445814 EFN89112.1 KQ976642 KYM78659.1 ADTU01011666 GL764129 EFZ18315.1 KQ981727 KYN37185.1 KQ977565 KYN01942.1 KQ982298 KYQ58161.1 GECU01030784 JAS76922.1 GEBQ01016844 JAT23133.1 KQ759828 OAD62665.1 LBMM01004406 KMQ92450.1 GL443983 EFN61406.1 KK107109 QOIP01000008 EZA59124.1 RLU19368.1 GALX01002864 JAB65602.1 GBYB01009518 JAG79285.1 BT127363 KB632025 AEE62325.1 ERL88094.1 APGK01029405 KB740734 ENN79256.1 GEZM01091886 JAV56723.1 NNAY01004010 OXU18500.1 KK852822 KDR15521.1 GDAI01001544 JAI16059.1 NEVH01020861 PNF20940.1 JRES01000960 KNC26729.1 KA647750 AFP62379.1 GANO01001608 JAB58263.1 CH478103 EAT33964.1 GAPW01003031 JAC10567.1 CH480824 EDW56559.1 CM000362 CM002911 EDX08002.1 KMY95425.1 AE013599 AY069084 CH954179 EDV55393.1 CM000158 EDW91312.1 CH902619 EDV36494.1 GDIQ01142444 LRGB01000141 JAL09282.1 KZS20632.1 GL732674 EFX67580.1 CP012524 ALC40499.1 GFDF01000185 JAV13899.1 CH964282 EDW85807.1 GL888463 EGI60606.1 GFDL01016217 JAV18828.1 JXJN01002473 AAAB01008960 EAA10831.2 CVRI01000001 CRK86190.1 ATLV01023245 KE525341 KFB48704.1 GDRN01147850 JAI56642.1 GDIQ01010478 JAN84259.1 DS235879 EEB19927.1 GAMC01002505 GAMC01002504 JAC04051.1 CH940648 EDW62149.1 LJIG01003010 KRT83930.1 GDHF01029648 GDHF01026689 GDHF01008611 JAI22666.1 JAI25625.1 JAI43703.1 AXCM01003909 GAKP01000670 GAKP01000669 JAC58282.1 AXCN02000522 GBXI01013099 GBXI01006354 JAD01193.1 JAD07938.1 GFWZ01000118 MBW20108.1 QCYY01003454 ROT63295.1 GALA01000970 JAA93882.1 CH479210 EDW32820.1 CM000071 EAL26487.1 JH432004 OUUW01000001 SPP74854.1 GAMD01003111 JAA98479.1 LJIJ01000045 ODN04389.1 GGFK01006130 MBW39451.1 KQ421056 KOF78522.1 GEFM01004463 JAP71333.1 IAAA01004788 LAA00496.1 GGFL01004069 MBW68247.1
JAT85083.1 ODYU01010630 SOQ55871.1 KZ150068 PZC74113.1 NWSH01002895 PCG67352.1 JTDY01000905 KOB75490.1 AGBW02008682 OWR52807.1 GAIX01003294 JAA89266.1 KQ971351 EFA06446.1 KQ414667 KOC64843.1 KQ434809 KZC06589.1 GL445814 EFN89112.1 KQ976642 KYM78659.1 ADTU01011666 GL764129 EFZ18315.1 KQ981727 KYN37185.1 KQ977565 KYN01942.1 KQ982298 KYQ58161.1 GECU01030784 JAS76922.1 GEBQ01016844 JAT23133.1 KQ759828 OAD62665.1 LBMM01004406 KMQ92450.1 GL443983 EFN61406.1 KK107109 QOIP01000008 EZA59124.1 RLU19368.1 GALX01002864 JAB65602.1 GBYB01009518 JAG79285.1 BT127363 KB632025 AEE62325.1 ERL88094.1 APGK01029405 KB740734 ENN79256.1 GEZM01091886 JAV56723.1 NNAY01004010 OXU18500.1 KK852822 KDR15521.1 GDAI01001544 JAI16059.1 NEVH01020861 PNF20940.1 JRES01000960 KNC26729.1 KA647750 AFP62379.1 GANO01001608 JAB58263.1 CH478103 EAT33964.1 GAPW01003031 JAC10567.1 CH480824 EDW56559.1 CM000362 CM002911 EDX08002.1 KMY95425.1 AE013599 AY069084 CH954179 EDV55393.1 CM000158 EDW91312.1 CH902619 EDV36494.1 GDIQ01142444 LRGB01000141 JAL09282.1 KZS20632.1 GL732674 EFX67580.1 CP012524 ALC40499.1 GFDF01000185 JAV13899.1 CH964282 EDW85807.1 GL888463 EGI60606.1 GFDL01016217 JAV18828.1 JXJN01002473 AAAB01008960 EAA10831.2 CVRI01000001 CRK86190.1 ATLV01023245 KE525341 KFB48704.1 GDRN01147850 JAI56642.1 GDIQ01010478 JAN84259.1 DS235879 EEB19927.1 GAMC01002505 GAMC01002504 JAC04051.1 CH940648 EDW62149.1 LJIG01003010 KRT83930.1 GDHF01029648 GDHF01026689 GDHF01008611 JAI22666.1 JAI25625.1 JAI43703.1 AXCM01003909 GAKP01000670 GAKP01000669 JAC58282.1 AXCN02000522 GBXI01013099 GBXI01006354 JAD01193.1 JAD07938.1 GFWZ01000118 MBW20108.1 QCYY01003454 ROT63295.1 GALA01000970 JAA93882.1 CH479210 EDW32820.1 CM000071 EAL26487.1 JH432004 OUUW01000001 SPP74854.1 GAMD01003111 JAA98479.1 LJIJ01000045 ODN04389.1 GGFK01006130 MBW39451.1 KQ421056 KOF78522.1 GEFM01004463 JAP71333.1 IAAA01004788 LAA00496.1 GGFL01004069 MBW68247.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000037510
UP000007151
+ More
UP000007266 UP000005203 UP000053825 UP000076502 UP000008237 UP000078540 UP000005205 UP000078541 UP000078542 UP000075809 UP000036403 UP000192223 UP000000311 UP000053097 UP000279307 UP000030742 UP000019118 UP000215335 UP000027135 UP000002358 UP000235965 UP000037069 UP000095301 UP000095300 UP000008820 UP000001292 UP000000304 UP000000803 UP000008711 UP000002282 UP000192221 UP000007801 UP000076858 UP000000305 UP000092553 UP000007798 UP000078200 UP000007755 UP000092460 UP000075885 UP000091820 UP000075903 UP000075882 UP000007062 UP000075900 UP000183832 UP000030765 UP000076408 UP000075920 UP000075880 UP000009046 UP000008792 UP000075883 UP000075886 UP000283509 UP000008744 UP000001819 UP000268350 UP000094527 UP000075881 UP000053454 UP000085678 UP000079169 UP000069272
UP000007266 UP000005203 UP000053825 UP000076502 UP000008237 UP000078540 UP000005205 UP000078541 UP000078542 UP000075809 UP000036403 UP000192223 UP000000311 UP000053097 UP000279307 UP000030742 UP000019118 UP000215335 UP000027135 UP000002358 UP000235965 UP000037069 UP000095301 UP000095300 UP000008820 UP000001292 UP000000304 UP000000803 UP000008711 UP000002282 UP000192221 UP000007801 UP000076858 UP000000305 UP000092553 UP000007798 UP000078200 UP000007755 UP000092460 UP000075885 UP000091820 UP000075903 UP000075882 UP000007062 UP000075900 UP000183832 UP000030765 UP000076408 UP000075920 UP000075880 UP000009046 UP000008792 UP000075883 UP000075886 UP000283509 UP000008744 UP000001819 UP000268350 UP000094527 UP000075881 UP000053454 UP000085678 UP000079169 UP000069272
Pfam
PF01267 F-actin_cap_A
Interpro
SUPFAM
SSF90096
SSF90096
Gene 3D
ProteinModelPortal
H9JT03
A0A194QWZ9
A0A194QF17
A0A1E1WDM6
A0A2H1WS37
A0A2W1BQV7
+ More
A0A2A4J7F6 A0A0L7LJ82 A0A212FGF2 S4PAE9 D6WRI1 A0A088A627 A0A0L7R1Z8 A0A154P3S0 E2B5B7 A0A151HZP1 A0A158NC62 E9IM12 A0A195F9A8 A0A195CN90 A0A151XCQ4 A0A1B6HQG9 A0A1B6LHE9 A0A310SN37 A0A0J7NIW0 A0A1W4XP49 E2AYP3 A0A026WVL7 V5GVA1 A0A0C9Q5X5 J3JVU5 N6UF55 A0A1Y1K5N3 A0A232EJF2 A0A067QYV3 A0A0K8TNW5 K7IR84 A0A2J7PX86 A0A0L0C5M5 T1PHZ2 A0A1I8P8L5 U5EU77 Q16I64 A0A023EMK3 B4I7I4 B4QFL0 Q9W2N0 B3NK27 B4P9J2 A0A1W4V5X5 B3MDV9 A0A0P5PYC3 E9HL07 A0A0M3QUE6 A0A1L8E5K2 B4NN68 A0A1A9V626 F4WYV2 A0A1Q3EUF1 A0A1B0ARK0 A0A182PDR9 A0A1A9WHV4 A0A182VAZ1 A0A182KNZ2 Q7Q5Y1 A0A182RQX4 A0A1J1HI91 A0A084WER0 A0A0P4VN78 A0A182Y2R4 A0A182WJX1 A0A0P6I4S5 A0A182IMQ9 E0W2S1 W8BRT2 B4LNG9 A0A0T6B9C2 A0A0K8UH13 A0A182MM38 A0A034WVC3 A0A182QSV4 A0A0A1WRX5 A0A2I9LP04 A0A3R7QCH9 T1E2U7 B4H4Y4 Q28X29 T1JNV6 A0A3B0J161 T1DFJ8 A0A1D2NGM3 A0A182K4R5 A0A2M4AF83 A0A0L8GNQ0 A0A131XVK7 A0A2L2XWT3 A0A1S3JR53 A0A1S3D469 A0A182FEI7 A0A2M4CSF0
A0A2A4J7F6 A0A0L7LJ82 A0A212FGF2 S4PAE9 D6WRI1 A0A088A627 A0A0L7R1Z8 A0A154P3S0 E2B5B7 A0A151HZP1 A0A158NC62 E9IM12 A0A195F9A8 A0A195CN90 A0A151XCQ4 A0A1B6HQG9 A0A1B6LHE9 A0A310SN37 A0A0J7NIW0 A0A1W4XP49 E2AYP3 A0A026WVL7 V5GVA1 A0A0C9Q5X5 J3JVU5 N6UF55 A0A1Y1K5N3 A0A232EJF2 A0A067QYV3 A0A0K8TNW5 K7IR84 A0A2J7PX86 A0A0L0C5M5 T1PHZ2 A0A1I8P8L5 U5EU77 Q16I64 A0A023EMK3 B4I7I4 B4QFL0 Q9W2N0 B3NK27 B4P9J2 A0A1W4V5X5 B3MDV9 A0A0P5PYC3 E9HL07 A0A0M3QUE6 A0A1L8E5K2 B4NN68 A0A1A9V626 F4WYV2 A0A1Q3EUF1 A0A1B0ARK0 A0A182PDR9 A0A1A9WHV4 A0A182VAZ1 A0A182KNZ2 Q7Q5Y1 A0A182RQX4 A0A1J1HI91 A0A084WER0 A0A0P4VN78 A0A182Y2R4 A0A182WJX1 A0A0P6I4S5 A0A182IMQ9 E0W2S1 W8BRT2 B4LNG9 A0A0T6B9C2 A0A0K8UH13 A0A182MM38 A0A034WVC3 A0A182QSV4 A0A0A1WRX5 A0A2I9LP04 A0A3R7QCH9 T1E2U7 B4H4Y4 Q28X29 T1JNV6 A0A3B0J161 T1DFJ8 A0A1D2NGM3 A0A182K4R5 A0A2M4AF83 A0A0L8GNQ0 A0A131XVK7 A0A2L2XWT3 A0A1S3JR53 A0A1S3D469 A0A182FEI7 A0A2M4CSF0
PDB
3LK4
E-value=4.02693e-97,
Score=904
Ontologies
GO
GO:0003779
GO:0008290
GO:0051016
GO:0051015
GO:0030479
GO:0030036
GO:0007015
GO:0016324
GO:0035220
GO:0005912
GO:0010591
GO:0046982
GO:0051490
GO:0046329
GO:0000902
GO:0007294
GO:0005869
GO:0007018
GO:0030837
GO:0051489
GO:0071203
GO:0032403
GO:0003676
GO:0016020
GO:0005515
GO:0006811
GO:0005622
GO:0006810
GO:0003824
PANTHER
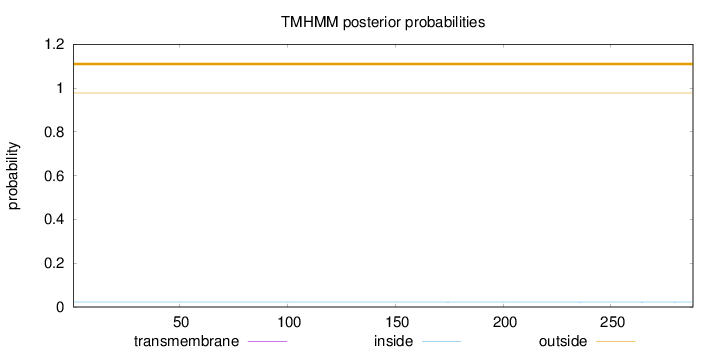
Topology
Length:
288
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00024
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02250
outside
1 - 288
Population Genetic Test Statistics
Pi
186.020888
Theta
197.419498
Tajima's D
-0.397055
CLR
19.786213
CSRT
0.261036948152592
Interpretation
Uncertain
