Gene
KWMTBOMO11201 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012632
Annotation
NADH-ubiquinone_oxidoreductase_subunit_B14.7_[Bombyx_mori]
Full name
Palmitoyltransferase
+ More
Tetraspanin
Tetraspanin
Location in the cell
Cytoplasmic Reliability : 1.487 PlasmaMembrane Reliability : 1.481
Sequence
CDS
ATGAATAAACTATTAAATTACAAATACTACGATACGCCTGAGGGCCAAGACATATTCATGAAGACATTTGTAACCAGTAAATATGCTGCAGTAGCCGGTCTTGCTGGTGCTTCTTTTGATGTGCTTATGTTCTCTCATCCAAAAGGTTTTGTCAATACAATCGGTCGAATGGGCTATATTGTAGGCCCTCTAGTGGGTATGGCAGTTGCCTTCACATTCACTACCAACGTTGCCCAAAATATTCGTGGGAAAAACGATAAACTCAATTACTTCTTGGGTGGTGCCACATCTGGCTTTGTGTTTAGTGCTTGGATGAAAAAGGGGATTATCGCGGTGCCTGCCGCGGTGGTACTAGGCGCAATAGCTGTGGTAAAGAAGACAGGCATCGATGAAGGCTGGATCTTTTTCCCAGATGCGGCACAATCTACTAAAACAATCAAATCAGTTAAACATGACTGGACTCTTCTCAAAGATATTGAAGAATTAAAGGGATGGACAGACGGTTCCAAGCAATAG
Protein
MNKLLNYKYYDTPEGQDIFMKTFVTSKYAAVAGLAGASFDVLMFSHPKGFVNTIGRMGYIVGPLVGMAVAFTFTTNVAQNIRGKNDKLNYFLGGATSGFVFSAWMKKGIIAVPAAVVLGAIAVVKKTGIDEGWIFFPDAAQSTKTIKSVKHDWTLLKDIEELKGWTDGSKQ
Summary
Catalytic Activity
hexadecanoyl-CoA + L-cysteinyl-[protein] = CoA + S-hexadecanoyl-L-cysteinyl-[protein]
Similarity
Belongs to the DHHC palmitoyltransferase family.
Belongs to the tetraspanin (TM4SF) family.
Belongs to the tetraspanin (TM4SF) family.
Uniprot
H9JSX0
Q2F5X5
A0A2H1WS21
A0A2W1BGB7
A0A2A4J7G8
A0A3S2NM38
+ More
I4DJX7 A0A194QF78 A0A194R0W6 S4P8X7 A0A212FGE8 A0A0L7LJV9 B4H4Y7 A0A1L8EBD7 A0A1L8EB13 A0A1I8P1W7 A0A034V176 A0A0K8U3I9 A0A0A1XFM5 A0A0L0C354 A0A2J7PGL6 A0A067RHJ5 Q28X30 W8BZR1 U5EST1 A0A2P8XEI1 A0A1B0AF86 B3NK25 A0A3B0JQB5 B4I7I2 B4QFK7 A0A1W4VIH0 T1E7Z3 B4P9J4 A0A2M4ACF4 A0A2M3Z6Y9 A0A1A9YKW4 A0A0K8TQN6 B4KT37 B4J8P7 A0A1J1J4P2 A0A2M4C1J2 A0A1W4X461 W5J8M2 Q7JYH3 A0A164TZ31 B4LQ78 A0A2M4C1K7 B4NN64 A0A0P4X751 A0A182FEI5 A0A182IQW0 A0A0P6I2F1 A0A182K3B8 A0A0M4EEL8 A0A182I2Q4 A0A182R3J5 A0A084WER2 A0A182YMK9 A0A182N925 T1D542 A0A182SVS9 A0A182XLI8 A0A182KQN7 Q7QJ20 A0A182PP42 A0A182QK50 Q1HQR1 A0A182VHM8 A0A182TX21 A0A023EGK5 A0A023EIL9 A0A182MKV2 A0A182WJS3 B0WVX9 A0A336MGC7 B3MDW1 A0A336M1L6 A0A0P4VJ48 R4FK60 A0A224XLT0 G3CJT7 A0A023F5Z7 A0A1I8MKN8 A0A2A4K455 A0A140DKV6 A0A0A9YBF5 A0A1L8E0C1 A0A1B0DDU4 A0A1Q3FKS7 A0A2A4K479 A0A069DPG4 A0A2H1WAF4 A0A0P5HMD9 A0A1B0CLN0 A0A2R7VRG7 A0A2H8TTV0 C4WY09
I4DJX7 A0A194QF78 A0A194R0W6 S4P8X7 A0A212FGE8 A0A0L7LJV9 B4H4Y7 A0A1L8EBD7 A0A1L8EB13 A0A1I8P1W7 A0A034V176 A0A0K8U3I9 A0A0A1XFM5 A0A0L0C354 A0A2J7PGL6 A0A067RHJ5 Q28X30 W8BZR1 U5EST1 A0A2P8XEI1 A0A1B0AF86 B3NK25 A0A3B0JQB5 B4I7I2 B4QFK7 A0A1W4VIH0 T1E7Z3 B4P9J4 A0A2M4ACF4 A0A2M3Z6Y9 A0A1A9YKW4 A0A0K8TQN6 B4KT37 B4J8P7 A0A1J1J4P2 A0A2M4C1J2 A0A1W4X461 W5J8M2 Q7JYH3 A0A164TZ31 B4LQ78 A0A2M4C1K7 B4NN64 A0A0P4X751 A0A182FEI5 A0A182IQW0 A0A0P6I2F1 A0A182K3B8 A0A0M4EEL8 A0A182I2Q4 A0A182R3J5 A0A084WER2 A0A182YMK9 A0A182N925 T1D542 A0A182SVS9 A0A182XLI8 A0A182KQN7 Q7QJ20 A0A182PP42 A0A182QK50 Q1HQR1 A0A182VHM8 A0A182TX21 A0A023EGK5 A0A023EIL9 A0A182MKV2 A0A182WJS3 B0WVX9 A0A336MGC7 B3MDW1 A0A336M1L6 A0A0P4VJ48 R4FK60 A0A224XLT0 G3CJT7 A0A023F5Z7 A0A1I8MKN8 A0A2A4K455 A0A140DKV6 A0A0A9YBF5 A0A1L8E0C1 A0A1B0DDU4 A0A1Q3FKS7 A0A2A4K479 A0A069DPG4 A0A2H1WAF4 A0A0P5HMD9 A0A1B0CLN0 A0A2R7VRG7 A0A2H8TTV0 C4WY09
EC Number
2.3.1.225
Pubmed
19121390
28756777
22651552
26354079
23622113
22118469
+ More
26227816 17994087 25348373 25830018 26108605 24845553 15632085 24495485 29403074 22936249 17550304 26369729 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 24438588 25244985 24330624 20966253 12364791 14747013 17210077 17204158 17510324 24945155 26483478 27129103 21058630 25474469 25315136 25401762 26823975 26334808
26227816 17994087 25348373 25830018 26108605 24845553 15632085 24495485 29403074 22936249 17550304 26369729 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 24438588 25244985 24330624 20966253 12364791 14747013 17210077 17204158 17510324 24945155 26483478 27129103 21058630 25474469 25315136 25401762 26823975 26334808
EMBL
BABH01038957
DQ311298
ABD36242.1
ODYU01010630
SOQ55870.1
KZ150068
+ More
PZC74112.1 NWSH01002895 PCG67353.1 RSAL01000270 RVE43188.1 AK401595 BAM18217.1 KQ459053 KPJ04137.1 KQ461073 KPJ09486.1 GAIX01003984 JAA88576.1 AGBW02008682 OWR52808.1 JTDY01000905 KOB75491.1 CH479210 EDW32823.1 GFDG01002883 JAV15916.1 GFDG01002878 JAV15921.1 GAKP01022713 JAC36239.1 GDHF01031414 JAI20900.1 GBXI01004530 JAD09762.1 JRES01000960 KNC26696.1 NEVH01025174 PNF15483.1 KK852680 KDR18623.1 CM000071 EAL26486.2 GAMC01001693 JAC04863.1 GANO01003112 JAB56759.1 PYGN01002482 PSN30416.1 CH954179 EDV55391.1 OUUW01000001 SPP74851.1 CH480824 EDW56557.1 CM000362 CM002911 EDX07999.1 KMY95423.1 GAMD01003208 JAA98382.1 CM000158 EDW91314.1 GGFK01005140 MBW38461.1 GGFM01003536 MBW24287.1 GDAI01001135 JAI16468.1 CH933808 EDW09557.1 CH916367 EDW01314.1 CVRI01000067 CRL06430.1 GGFJ01010049 MBW59190.1 ADMH02002097 ETN59225.1 AY070603 AY075551 AE013599 AAL48074.1 AAL68358.1 AAM68190.1 LRGB01001663 KZS10901.1 CH940648 EDW61363.1 KRF79955.1 GGFJ01010046 MBW59187.1 CH964282 EDW85803.1 GDIP01246818 JAI76583.1 GDIQ01010631 JAN84106.1 CP012524 ALC40497.1 APCN01000178 ATLV01023246 KE525341 KFB48706.1 GALA01000693 JAA94159.1 AAAB01008807 EAA04349.2 AXCN02000540 DQ440383 CH478132 ABF18416.1 EAT33861.1 GAPW01005140 JAC08458.1 JXUM01035232 JXUM01035233 GAPW01005139 KQ561052 JAC08459.1 KXJ79870.1 AXCM01010371 DS232134 EDS35811.1 UFQT01001198 SSX29442.1 CH902619 EDV36496.1 UFQT01000322 SSX23231.1 GDKW01001626 JAI54969.1 ACPB03011824 GAHY01002013 JAA75497.1 GFTR01002980 JAW13446.1 HP639854 AEM97991.1 GBBI01002089 JAC16623.1 NWSH01000149 PCG79035.1 KT748790 AMK48571.1 GBHO01016739 GBRD01014295 GDHC01001756 JAG26865.1 JAG51531.1 JAQ16873.1 GFDF01001947 JAV12137.1 AJVK01032284 GFDL01006910 JAV28135.1 PCG79057.1 GBGD01003164 JAC85725.1 ODYU01007345 SOQ50030.1 GDIQ01226068 JAK25657.1 AJWK01017457 KK854010 PTY09260.1 GFXV01005574 GFXV01007558 MBW17379.1 MBW19363.1 ABLF02037656 AK342962 BAH72779.1
PZC74112.1 NWSH01002895 PCG67353.1 RSAL01000270 RVE43188.1 AK401595 BAM18217.1 KQ459053 KPJ04137.1 KQ461073 KPJ09486.1 GAIX01003984 JAA88576.1 AGBW02008682 OWR52808.1 JTDY01000905 KOB75491.1 CH479210 EDW32823.1 GFDG01002883 JAV15916.1 GFDG01002878 JAV15921.1 GAKP01022713 JAC36239.1 GDHF01031414 JAI20900.1 GBXI01004530 JAD09762.1 JRES01000960 KNC26696.1 NEVH01025174 PNF15483.1 KK852680 KDR18623.1 CM000071 EAL26486.2 GAMC01001693 JAC04863.1 GANO01003112 JAB56759.1 PYGN01002482 PSN30416.1 CH954179 EDV55391.1 OUUW01000001 SPP74851.1 CH480824 EDW56557.1 CM000362 CM002911 EDX07999.1 KMY95423.1 GAMD01003208 JAA98382.1 CM000158 EDW91314.1 GGFK01005140 MBW38461.1 GGFM01003536 MBW24287.1 GDAI01001135 JAI16468.1 CH933808 EDW09557.1 CH916367 EDW01314.1 CVRI01000067 CRL06430.1 GGFJ01010049 MBW59190.1 ADMH02002097 ETN59225.1 AY070603 AY075551 AE013599 AAL48074.1 AAL68358.1 AAM68190.1 LRGB01001663 KZS10901.1 CH940648 EDW61363.1 KRF79955.1 GGFJ01010046 MBW59187.1 CH964282 EDW85803.1 GDIP01246818 JAI76583.1 GDIQ01010631 JAN84106.1 CP012524 ALC40497.1 APCN01000178 ATLV01023246 KE525341 KFB48706.1 GALA01000693 JAA94159.1 AAAB01008807 EAA04349.2 AXCN02000540 DQ440383 CH478132 ABF18416.1 EAT33861.1 GAPW01005140 JAC08458.1 JXUM01035232 JXUM01035233 GAPW01005139 KQ561052 JAC08459.1 KXJ79870.1 AXCM01010371 DS232134 EDS35811.1 UFQT01001198 SSX29442.1 CH902619 EDV36496.1 UFQT01000322 SSX23231.1 GDKW01001626 JAI54969.1 ACPB03011824 GAHY01002013 JAA75497.1 GFTR01002980 JAW13446.1 HP639854 AEM97991.1 GBBI01002089 JAC16623.1 NWSH01000149 PCG79035.1 KT748790 AMK48571.1 GBHO01016739 GBRD01014295 GDHC01001756 JAG26865.1 JAG51531.1 JAQ16873.1 GFDF01001947 JAV12137.1 AJVK01032284 GFDL01006910 JAV28135.1 PCG79057.1 GBGD01003164 JAC85725.1 ODYU01007345 SOQ50030.1 GDIQ01226068 JAK25657.1 AJWK01017457 KK854010 PTY09260.1 GFXV01005574 GFXV01007558 MBW17379.1 MBW19363.1 ABLF02037656 AK342962 BAH72779.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000007151
+ More
UP000037510 UP000008744 UP000095300 UP000037069 UP000235965 UP000027135 UP000001819 UP000245037 UP000092445 UP000008711 UP000268350 UP000001292 UP000000304 UP000192221 UP000002282 UP000092443 UP000009192 UP000001070 UP000183832 UP000192223 UP000000673 UP000000803 UP000076858 UP000008792 UP000007798 UP000069272 UP000075880 UP000075881 UP000092553 UP000075840 UP000075900 UP000030765 UP000076408 UP000075884 UP000075901 UP000076407 UP000075882 UP000007062 UP000075885 UP000075886 UP000008820 UP000075903 UP000075902 UP000069940 UP000249989 UP000075883 UP000075920 UP000002320 UP000007801 UP000015103 UP000095301 UP000092462 UP000092461 UP000007819
UP000037510 UP000008744 UP000095300 UP000037069 UP000235965 UP000027135 UP000001819 UP000245037 UP000092445 UP000008711 UP000268350 UP000001292 UP000000304 UP000192221 UP000002282 UP000092443 UP000009192 UP000001070 UP000183832 UP000192223 UP000000673 UP000000803 UP000076858 UP000008792 UP000007798 UP000069272 UP000075880 UP000075881 UP000092553 UP000075840 UP000075900 UP000030765 UP000076408 UP000075884 UP000075901 UP000076407 UP000075882 UP000007062 UP000075885 UP000075886 UP000008820 UP000075903 UP000075902 UP000069940 UP000249989 UP000075883 UP000075920 UP000002320 UP000007801 UP000015103 UP000095301 UP000092462 UP000092461 UP000007819
Interpro
SUPFAM
SSF48652
SSF48652
Gene 3D
ProteinModelPortal
H9JSX0
Q2F5X5
A0A2H1WS21
A0A2W1BGB7
A0A2A4J7G8
A0A3S2NM38
+ More
I4DJX7 A0A194QF78 A0A194R0W6 S4P8X7 A0A212FGE8 A0A0L7LJV9 B4H4Y7 A0A1L8EBD7 A0A1L8EB13 A0A1I8P1W7 A0A034V176 A0A0K8U3I9 A0A0A1XFM5 A0A0L0C354 A0A2J7PGL6 A0A067RHJ5 Q28X30 W8BZR1 U5EST1 A0A2P8XEI1 A0A1B0AF86 B3NK25 A0A3B0JQB5 B4I7I2 B4QFK7 A0A1W4VIH0 T1E7Z3 B4P9J4 A0A2M4ACF4 A0A2M3Z6Y9 A0A1A9YKW4 A0A0K8TQN6 B4KT37 B4J8P7 A0A1J1J4P2 A0A2M4C1J2 A0A1W4X461 W5J8M2 Q7JYH3 A0A164TZ31 B4LQ78 A0A2M4C1K7 B4NN64 A0A0P4X751 A0A182FEI5 A0A182IQW0 A0A0P6I2F1 A0A182K3B8 A0A0M4EEL8 A0A182I2Q4 A0A182R3J5 A0A084WER2 A0A182YMK9 A0A182N925 T1D542 A0A182SVS9 A0A182XLI8 A0A182KQN7 Q7QJ20 A0A182PP42 A0A182QK50 Q1HQR1 A0A182VHM8 A0A182TX21 A0A023EGK5 A0A023EIL9 A0A182MKV2 A0A182WJS3 B0WVX9 A0A336MGC7 B3MDW1 A0A336M1L6 A0A0P4VJ48 R4FK60 A0A224XLT0 G3CJT7 A0A023F5Z7 A0A1I8MKN8 A0A2A4K455 A0A140DKV6 A0A0A9YBF5 A0A1L8E0C1 A0A1B0DDU4 A0A1Q3FKS7 A0A2A4K479 A0A069DPG4 A0A2H1WAF4 A0A0P5HMD9 A0A1B0CLN0 A0A2R7VRG7 A0A2H8TTV0 C4WY09
I4DJX7 A0A194QF78 A0A194R0W6 S4P8X7 A0A212FGE8 A0A0L7LJV9 B4H4Y7 A0A1L8EBD7 A0A1L8EB13 A0A1I8P1W7 A0A034V176 A0A0K8U3I9 A0A0A1XFM5 A0A0L0C354 A0A2J7PGL6 A0A067RHJ5 Q28X30 W8BZR1 U5EST1 A0A2P8XEI1 A0A1B0AF86 B3NK25 A0A3B0JQB5 B4I7I2 B4QFK7 A0A1W4VIH0 T1E7Z3 B4P9J4 A0A2M4ACF4 A0A2M3Z6Y9 A0A1A9YKW4 A0A0K8TQN6 B4KT37 B4J8P7 A0A1J1J4P2 A0A2M4C1J2 A0A1W4X461 W5J8M2 Q7JYH3 A0A164TZ31 B4LQ78 A0A2M4C1K7 B4NN64 A0A0P4X751 A0A182FEI5 A0A182IQW0 A0A0P6I2F1 A0A182K3B8 A0A0M4EEL8 A0A182I2Q4 A0A182R3J5 A0A084WER2 A0A182YMK9 A0A182N925 T1D542 A0A182SVS9 A0A182XLI8 A0A182KQN7 Q7QJ20 A0A182PP42 A0A182QK50 Q1HQR1 A0A182VHM8 A0A182TX21 A0A023EGK5 A0A023EIL9 A0A182MKV2 A0A182WJS3 B0WVX9 A0A336MGC7 B3MDW1 A0A336M1L6 A0A0P4VJ48 R4FK60 A0A224XLT0 G3CJT7 A0A023F5Z7 A0A1I8MKN8 A0A2A4K455 A0A140DKV6 A0A0A9YBF5 A0A1L8E0C1 A0A1B0DDU4 A0A1Q3FKS7 A0A2A4K479 A0A069DPG4 A0A2H1WAF4 A0A0P5HMD9 A0A1B0CLN0 A0A2R7VRG7 A0A2H8TTV0 C4WY09
PDB
5XTI
E-value=1.59471e-12,
Score=171
Ontologies
PATHWAY
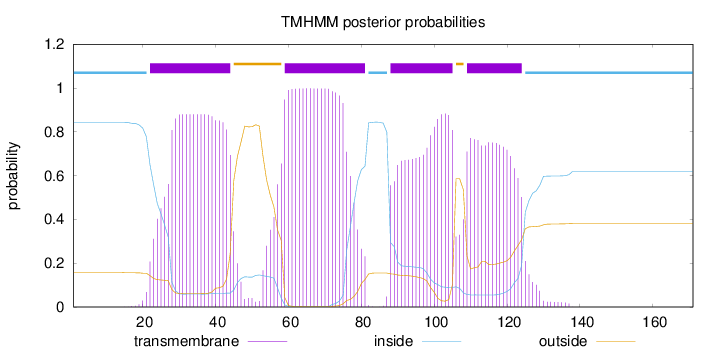
Topology
Subcellular location
Membrane
Length:
171
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
66.15551
Exp number, first 60 AAs:
22.25338
Total prob of N-in:
0.84237
POSSIBLE N-term signal
sequence
inside
1 - 21
TMhelix
22 - 44
outside
45 - 58
TMhelix
59 - 81
inside
82 - 87
TMhelix
88 - 105
outside
106 - 108
TMhelix
109 - 124
inside
125 - 171
Population Genetic Test Statistics
Pi
177.883462
Theta
184.398152
Tajima's D
-0.410093
CLR
0.189014
CSRT
0.262986850657467
Interpretation
Uncertain
