Pre Gene Modal
BGIBMGA012662
Annotation
PREDICTED:_focadhesin_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 1.764
Sequence
CDS
ATGGATATTGAGAACCGAGGACCGGGTCTAGAACTCCTTAGAGCAGTCTTCCTCTGGTTGACGTGTAACCCTCAACGAGACGGGGGCTCGAGGCCGTGGCAGCTGTTGCTGAGTCTCCCCCAGGGTCAAGCTAAGTCGTTGCTGCTCCTAGCTTGCCTCAGTTGCCAACAGATATGCAACTCGTCGTTAATAGAGAGAGCATTCAGTGCTTACTCGGCTGTCACCGATGCCGCCATTTACCAGCAGGATAAAGAGACGGTTAGGGCCTTACTGCCCATGCTGGCTAGAATCACCAATGAGCTGATCAAGCATGGTCGTGACCCGCGTCCCTGCTACATCCTGATGGAGCGTTGCTTCAACCTGGACGCTGCGGAACTCAGGACCGTCGCCGGCCTGGTTCTGATGATCTTAGCTGACAACTTGCCCCACACGTCGGCTCTGTACCTCCATGAGCTTTTTAACTTGTGTTTAAATATTATAAATAAATACGCGTACACACAGATCTCGATCAACTACCTGATCGCTTTAACGTTACAGTGGCTTAATATGCCATCTTATCTCACCGCTTCTGCCTTGAAAAGTGCATCGAGAATCCTCGATATTTACCAAGAGGACGCCGGAGAGGACACCAGGATGTACATGACGAATTTAAATACGAATAAAACATTTCAAACATTACTGCACACGGACAAACATCTTTCAATAGTCTACAATTTGAATCGAAACTATGAACGTCTGCGCGACAATCCGGACAAATTAAAATGTTGGGTGTCTGATATTAGTTCTCTAGATAACGCGGTCAAATACGACATGTTGCCGCTATTGCTCGGTGTTGCCATGTCGCAAAACATCGAGGGGGATCAGGAGGAGATCGTGTTGCAATCATTAAATATTTTAATTGATCTGGTTGATTATAACAAGGATATGTCTGTACAACTTTTACCGATCTTGCTCTATAAAATATCGCACGATATATCGCCGTCTATCAAAATGGAGTGTCTGAAGGCTTTACCGCTAATGGCTAAGAGCAAGGAAAATGTGCCAACAATAGTGTCGATATTGAACAAGCTGAAGGTCAATAAAGGTGTGCCGATTTCGTTCCTGATCAGTCTGTATACGAGTCTAAGTGAAACGCAGGTCCGCTGCTTTCCATACCTTCAGGAGTTGTTGGTTGACCCCGGCGCGGGGAGGCCAGACGATCTGAAGTGGGAAGTCGACGTAGCTAGAGCTCTATCGGTCAAACGCATCTGCGAGATAAGGGGCTCGTCTCACGGCGTGGAACTGGTGTCGGTGATCTCGACGCTGTTGAACCGCTGCGGCGACAAGTCCGGGTCGGCGGCGACCTCGGCCGCGCTGCAGAGCATCGCCCGGCTGTGGCGCAGCGCGTCCATTGCGCCGCCGAGCACGTGGCGCGCATTGGAACCCAAACTCGGACGCGACACACGACCTCTGGTGCAAATCAGCGTTTGTAACCTGCTTGCCGAAGTGCCAGCCCTGAGGGTGTCGACTCCTGACTACGACTGGCTGATCAGTGCCTCCTGCCGCCGGCTCTGGGCTTACGTGGCCGATTCGAATCATCCTGAAGTGGTGGAGGCAGCATGCGATGCCTTGGGTGGATATCTGTTGGATGATTACAAGCTTAAGGACATACCAGAGATCTACAGACGCACAGTCAAACTCCCTGCCTCGTACTGCAAGACTCCAGCTGACGCCGCCAGGAATCCTGAAGACGTTCTCGAGTATATTCCGTGCGAGGTGTGGCCGGAAGTGTTCAAGTGCAGTAACCAGGCGGCCCTGGGCGGAGTCGCGCGGCTGACGACCAGGCTGATCGAGCGCGAGATCAAGGGGTACAGGAGCGGCGTGTACCATTTGGAAGCCAAAACTGAGCCCATGGGCTACAACCACCTCCAGCCGTTCAGCGTCATCAGGGGCCTGATGGAATGTTTCAGGAAGCAGGCAACCTCACCCTCGTTCGACTACTCGGACGAGATCCTCCTGGCGATACTGCAGGCCCTGACCTCCGACTACCCCCGACCGCTGCCCCCCATAGACCTGTGCTTCCTGCCCGAAATGTTCCACCGCGGCCGCCAGTGGAAGGAACTGTCCGTAAGACTGGCCGCCCGGCAGGCTCTGGTCTCCATCTCTGCCAAGAGGATCATGGATAACTATCTGCAGAGTATCGAGGTGGAGAACTGTGATGAATCTGATATCCTGACAGTTTATGAACTCCTGCCGACCCTGTGCCGAGGGATGCCGCCGAACTCCCTTCGAGCTCCCATCGAGAAGACCCTAGGCAGGTCCCACGCTATCGTCTCAAAGACCAAGTCCTCGGACAATCCAGAGGAGCTGCTGTTCGTGAAACTGCTGGTGCTCATCCGGAGATGCTTGGAATCGGAGAAGATACACGACGCCAACAGGAAACTGCTCTCTCAGATGGTCGAGAATTATTATTCTGTTATTGACGAATCAAATGTGTCGTGGGACACATATGTAGAGACGTGCCGAAGTCTATCGTCAGCTCACCTGGAGCGCATGACGTCGCCCTCGTCGTGGTGGGAGGCCAGCGGCGAGGCCTTGAGGCGGGCGTGGCGCGTGAGGTGTGAGGTCGCGCGGGGGGCGCCCGGGGGGATCGCGCTCGTGTGGCTCAACGAGATGATCGACGCTCAAGCTTCCAGTCCACCCGAGCAAGACTTCTCCCTGCGCTGCATGGCGCCGGTACTGCAGGCGGCAGACCCTGGCTCCGCGGCCGCGAGGGACTGGTTCCTGCAGCTGATGGCGCGCACGCAGGTCGCCTTCAATGAAACGGAAGACTTGTCGGCCAAGCTGTACCTGTGCGACGTGTTCGTGCTGAGCGTGGTGACGCTGAGCGGGCACGGCGCGCTGCGGCCCGGGGACGCAGTCGCCGGGGACCGCGCCGCCGCCCGCGCGCTCTTCCCGGCCGCCGTCGCAGCGCTGTTCAGCCGAGAGCCCTGGGAACAGTCTACGCCACAGATCCTGGAATGGTTGTGTCACACCCGGAGCTCTGTGGCGGACCCCCGCATGGCGCAGTGCTGTCAGCGCGGCCTGCTGTCCATGCGGCACGCGGCTCACTTCGCGACGCACAACACTTGGATCAAATTAGAAAGTCACCTCGCCAGAGAAAACATTGAAGACTGA
Protein
MDIENRGPGLELLRAVFLWLTCNPQRDGGSRPWQLLLSLPQGQAKSLLLLACLSCQQICNSSLIERAFSAYSAVTDAAIYQQDKETVRALLPMLARITNELIKHGRDPRPCYILMERCFNLDAAELRTVAGLVLMILADNLPHTSALYLHELFNLCLNIINKYAYTQISINYLIALTLQWLNMPSYLTASALKSASRILDIYQEDAGEDTRMYMTNLNTNKTFQTLLHTDKHLSIVYNLNRNYERLRDNPDKLKCWVSDISSLDNAVKYDMLPLLLGVAMSQNIEGDQEEIVLQSLNILIDLVDYNKDMSVQLLPILLYKISHDISPSIKMECLKALPLMAKSKENVPTIVSILNKLKVNKGVPISFLISLYTSLSETQVRCFPYLQELLVDPGAGRPDDLKWEVDVARALSVKRICEIRGSSHGVELVSVISTLLNRCGDKSGSAATSAALQSIARLWRSASIAPPSTWRALEPKLGRDTRPLVQISVCNLLAEVPALRVSTPDYDWLISASCRRLWAYVADSNHPEVVEAACDALGGYLLDDYKLKDIPEIYRRTVKLPASYCKTPADAARNPEDVLEYIPCEVWPEVFKCSNQAALGGVARLTTRLIEREIKGYRSGVYHLEAKTEPMGYNHLQPFSVIRGLMECFRKQATSPSFDYSDEILLAILQALTSDYPRPLPPIDLCFLPEMFHRGRQWKELSVRLAARQALVSISAKRIMDNYLQSIEVENCDESDILTVYELLPTLCRGMPPNSLRAPIEKTLGRSHAIVSKTKSSDNPEELLFVKLLVLIRRCLESEKIHDANRKLLSQMVENYYSVIDESNVSWDTYVETCRSLSSAHLERMTSPSSWWEASGEALRRAWRVRCEVARGAPGGIALVWLNEMIDAQASSPPEQDFSLRCMAPVLQAADPGSAAARDWFLQLMARTQVAFNETEDLSAKLYLCDVFVLSVVTLSGHGALRPGDAVAGDRAAARALFPAAVAALFSREPWEQSTPQILEWLCHTRSSVADPRMAQCCQRGLLSMRHAAHFATHNTWIKLESHLARENIED
Summary
Uniprot
A0A2A4JVU3
A0A2H1W092
A0A194QFE4
A0A194QX04
A0A212ELX0
A0A067QTM1
+ More
A0A2J7Q7Z6 A0A2J7Q807 A0A2J7Q7Z1 A0A0L7QXL7 A0A2A3EAI8 A0A088A308 A0A0M9A7H6 E2AZD0 A0A310SGJ3 A0A026WSR1 A0A3L8D8I8 F4WG80 A0A158NT53 A0A154P5F4 A0A195EFH9 A0A151WJE6 A0A195EZR6 A0A195B6B8 A0A151IAK8 A0A0K8VSR4 A0A0K8W2L2 A0A0K8USI9 A0A0K8V3W1 A0A0A1WNI7 A0A0K8TWV5 E2BVS2 A0A1Y1MEH0 A0A0J7KZ56 A0A1B0FPN6 A0A1A9W9M3 A0A1A9VST1 A0A1B0AAY7 A0A1I8NY12 A0A0L0CBA8 A0A1B0BS51 A0A1A9YGH7 A0A1I8N8B6 T1PHB9 A0A034VIG4 A0A0P4VWJ2 D6WTF9 A0A0T6BGP8 B4LMB8 B4J7D8 A0A0Q9X988 B4KSF1 B4GHN8 A0A0M3QV37 Q28ZJ5 B3MF07 A0A3B0JMI2 A0A2P2I2Q4 A0A0C9QBD0 Q8MQP1 A0A1W4UM00 A0A0K8W0Z1 Q9W1Q5 A0A182YQ03 A0A0K8W5U7 B4P9L4 B4I8L9 A0A0J9RJQ7 A0A1B6HFK3 A0A182K8Y7 B3NPD7 B4NMN4 A0A182R082 A0A1S4F3N2 A0A182PGE7 A0A182HMG2 Q7Q7F2 A0A182KW16 Q0IG60 A0A182R2K5 B0VZ36 A0A182XD31 A0A1Q3F5H2 A0A182WAK8 A0A182LU14 A0A182UUC2 E0VQU2 A0A182U189 A0A182GLL5 A0A182GE88 A0A1W4XLG9 A0A2M4CP47
A0A2J7Q7Z6 A0A2J7Q807 A0A2J7Q7Z1 A0A0L7QXL7 A0A2A3EAI8 A0A088A308 A0A0M9A7H6 E2AZD0 A0A310SGJ3 A0A026WSR1 A0A3L8D8I8 F4WG80 A0A158NT53 A0A154P5F4 A0A195EFH9 A0A151WJE6 A0A195EZR6 A0A195B6B8 A0A151IAK8 A0A0K8VSR4 A0A0K8W2L2 A0A0K8USI9 A0A0K8V3W1 A0A0A1WNI7 A0A0K8TWV5 E2BVS2 A0A1Y1MEH0 A0A0J7KZ56 A0A1B0FPN6 A0A1A9W9M3 A0A1A9VST1 A0A1B0AAY7 A0A1I8NY12 A0A0L0CBA8 A0A1B0BS51 A0A1A9YGH7 A0A1I8N8B6 T1PHB9 A0A034VIG4 A0A0P4VWJ2 D6WTF9 A0A0T6BGP8 B4LMB8 B4J7D8 A0A0Q9X988 B4KSF1 B4GHN8 A0A0M3QV37 Q28ZJ5 B3MF07 A0A3B0JMI2 A0A2P2I2Q4 A0A0C9QBD0 Q8MQP1 A0A1W4UM00 A0A0K8W0Z1 Q9W1Q5 A0A182YQ03 A0A0K8W5U7 B4P9L4 B4I8L9 A0A0J9RJQ7 A0A1B6HFK3 A0A182K8Y7 B3NPD7 B4NMN4 A0A182R082 A0A1S4F3N2 A0A182PGE7 A0A182HMG2 Q7Q7F2 A0A182KW16 Q0IG60 A0A182R2K5 B0VZ36 A0A182XD31 A0A1Q3F5H2 A0A182WAK8 A0A182LU14 A0A182UUC2 E0VQU2 A0A182U189 A0A182GLL5 A0A182GE88 A0A1W4XLG9 A0A2M4CP47
Pubmed
26354079
22118469
24845553
20798317
24508170
30249741
+ More
21719571 21347285 25830018 28004739 26108605 25315136 25348373 18362917 19820115 17994087 18057021 15632085 23185243 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25244985 17550304 22936249 12364791 14747013 17210077 20966253 17510324 20566863 26483478
21719571 21347285 25830018 28004739 26108605 25315136 25348373 18362917 19820115 17994087 18057021 15632085 23185243 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25244985 17550304 22936249 12364791 14747013 17210077 20966253 17510324 20566863 26483478
EMBL
NWSH01000536
PCG75808.1
ODYU01005556
SOQ46520.1
KQ459053
KPJ04134.1
+ More
KQ461073 KPJ09490.1 AGBW02013975 OWR42469.1 KK852949 KDR13366.1 NEVH01016993 PNF24702.1 PNF24705.1 PNF24704.1 KQ414704 KOC63321.1 KZ288322 PBC28206.1 KQ435719 KOX78797.1 GL444207 EFN61169.1 KQ770178 OAD52680.1 KK107111 EZA59023.1 QOIP01000011 RLU16830.1 GL888128 EGI66847.1 ADTU01025540 KQ434809 KZC06418.1 KQ978957 KYN27030.1 KQ983039 KYQ47951.1 KQ981897 KYN33718.1 KQ976579 KYM80051.1 KQ978192 KYM96532.1 GDHF01010383 JAI41931.1 GDHF01007013 JAI45301.1 GDHF01022655 JAI29659.1 GDHF01018816 JAI33498.1 GBXI01014314 JAC99977.1 GDHF01033573 JAI18741.1 GL450957 EFN80200.1 GEZM01033997 JAV83991.1 LBMM01001720 KMQ95847.1 CCAG010023233 JRES01000655 KNC29505.1 JXJN01019518 KA648166 AFP62795.1 GAKP01016688 JAC42264.1 GDRN01102852 JAI58167.1 KQ971352 EFA06297.1 LJIG01000451 KRT86489.1 CH940648 EDW62013.1 KRF80384.1 KRF80385.1 KRF80386.1 CH916367 EDW01062.1 CH933808 KRG04677.1 EDW09456.1 CH479183 EDW36008.1 CP012524 ALC41703.1 CM000071 EAL25618.1 KRT02416.1 KRT02417.1 KRT02418.1 CH902619 EDV36628.1 OUUW01000001 SPP74769.1 IACF01002669 LAB68314.1 GBYB01000554 JAG70321.1 AY128471 AAM75064.1 GDHF01032825 GDHF01007502 JAI19489.1 JAI44812.1 AE013599 BT150465 AAF47000.2 AJP62087.1 GDHF01005877 JAI46437.1 CM000158 EDW92322.1 CH480824 EDW56944.1 CM002911 KMY96027.1 GECU01034315 JAS73391.1 CH954179 EDV56800.1 CH964282 EDW85623.1 AXCN02000905 AAAB01008960 EAA11021.3 CH477261 EAT45697.1 DS231813 EDS25627.1 GFDL01012239 JAV22806.1 AXCM01003851 DS235442 EEB15748.1 JXUM01013755 JXUM01013756 JXUM01013757 JXUM01013758 JXUM01013759 KQ560384 KXJ82677.1 JXUM01010147 KQ560300 KXJ83169.1 GGFL01002916 MBW67094.1
KQ461073 KPJ09490.1 AGBW02013975 OWR42469.1 KK852949 KDR13366.1 NEVH01016993 PNF24702.1 PNF24705.1 PNF24704.1 KQ414704 KOC63321.1 KZ288322 PBC28206.1 KQ435719 KOX78797.1 GL444207 EFN61169.1 KQ770178 OAD52680.1 KK107111 EZA59023.1 QOIP01000011 RLU16830.1 GL888128 EGI66847.1 ADTU01025540 KQ434809 KZC06418.1 KQ978957 KYN27030.1 KQ983039 KYQ47951.1 KQ981897 KYN33718.1 KQ976579 KYM80051.1 KQ978192 KYM96532.1 GDHF01010383 JAI41931.1 GDHF01007013 JAI45301.1 GDHF01022655 JAI29659.1 GDHF01018816 JAI33498.1 GBXI01014314 JAC99977.1 GDHF01033573 JAI18741.1 GL450957 EFN80200.1 GEZM01033997 JAV83991.1 LBMM01001720 KMQ95847.1 CCAG010023233 JRES01000655 KNC29505.1 JXJN01019518 KA648166 AFP62795.1 GAKP01016688 JAC42264.1 GDRN01102852 JAI58167.1 KQ971352 EFA06297.1 LJIG01000451 KRT86489.1 CH940648 EDW62013.1 KRF80384.1 KRF80385.1 KRF80386.1 CH916367 EDW01062.1 CH933808 KRG04677.1 EDW09456.1 CH479183 EDW36008.1 CP012524 ALC41703.1 CM000071 EAL25618.1 KRT02416.1 KRT02417.1 KRT02418.1 CH902619 EDV36628.1 OUUW01000001 SPP74769.1 IACF01002669 LAB68314.1 GBYB01000554 JAG70321.1 AY128471 AAM75064.1 GDHF01032825 GDHF01007502 JAI19489.1 JAI44812.1 AE013599 BT150465 AAF47000.2 AJP62087.1 GDHF01005877 JAI46437.1 CM000158 EDW92322.1 CH480824 EDW56944.1 CM002911 KMY96027.1 GECU01034315 JAS73391.1 CH954179 EDV56800.1 CH964282 EDW85623.1 AXCN02000905 AAAB01008960 EAA11021.3 CH477261 EAT45697.1 DS231813 EDS25627.1 GFDL01012239 JAV22806.1 AXCM01003851 DS235442 EEB15748.1 JXUM01013755 JXUM01013756 JXUM01013757 JXUM01013758 JXUM01013759 KQ560384 KXJ82677.1 JXUM01010147 KQ560300 KXJ83169.1 GGFL01002916 MBW67094.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000007151
UP000027135
UP000235965
+ More
UP000053825 UP000242457 UP000005203 UP000053105 UP000000311 UP000053097 UP000279307 UP000007755 UP000005205 UP000076502 UP000078492 UP000075809 UP000078541 UP000078540 UP000078542 UP000008237 UP000036403 UP000092444 UP000091820 UP000078200 UP000092445 UP000095300 UP000037069 UP000092460 UP000092443 UP000095301 UP000007266 UP000008792 UP000001070 UP000009192 UP000008744 UP000092553 UP000001819 UP000007801 UP000268350 UP000192221 UP000000803 UP000076408 UP000002282 UP000001292 UP000075881 UP000008711 UP000007798 UP000075886 UP000075885 UP000007062 UP000075882 UP000008820 UP000075900 UP000002320 UP000076407 UP000075920 UP000075883 UP000009046 UP000075902 UP000069940 UP000249989 UP000192223
UP000053825 UP000242457 UP000005203 UP000053105 UP000000311 UP000053097 UP000279307 UP000007755 UP000005205 UP000076502 UP000078492 UP000075809 UP000078541 UP000078540 UP000078542 UP000008237 UP000036403 UP000092444 UP000091820 UP000078200 UP000092445 UP000095300 UP000037069 UP000092460 UP000092443 UP000095301 UP000007266 UP000008792 UP000001070 UP000009192 UP000008744 UP000092553 UP000001819 UP000007801 UP000268350 UP000192221 UP000000803 UP000076408 UP000002282 UP000001292 UP000075881 UP000008711 UP000007798 UP000075886 UP000075885 UP000007062 UP000075882 UP000008820 UP000075900 UP000002320 UP000076407 UP000075920 UP000075883 UP000009046 UP000075902 UP000069940 UP000249989 UP000192223
Interpro
ProteinModelPortal
A0A2A4JVU3
A0A2H1W092
A0A194QFE4
A0A194QX04
A0A212ELX0
A0A067QTM1
+ More
A0A2J7Q7Z6 A0A2J7Q807 A0A2J7Q7Z1 A0A0L7QXL7 A0A2A3EAI8 A0A088A308 A0A0M9A7H6 E2AZD0 A0A310SGJ3 A0A026WSR1 A0A3L8D8I8 F4WG80 A0A158NT53 A0A154P5F4 A0A195EFH9 A0A151WJE6 A0A195EZR6 A0A195B6B8 A0A151IAK8 A0A0K8VSR4 A0A0K8W2L2 A0A0K8USI9 A0A0K8V3W1 A0A0A1WNI7 A0A0K8TWV5 E2BVS2 A0A1Y1MEH0 A0A0J7KZ56 A0A1B0FPN6 A0A1A9W9M3 A0A1A9VST1 A0A1B0AAY7 A0A1I8NY12 A0A0L0CBA8 A0A1B0BS51 A0A1A9YGH7 A0A1I8N8B6 T1PHB9 A0A034VIG4 A0A0P4VWJ2 D6WTF9 A0A0T6BGP8 B4LMB8 B4J7D8 A0A0Q9X988 B4KSF1 B4GHN8 A0A0M3QV37 Q28ZJ5 B3MF07 A0A3B0JMI2 A0A2P2I2Q4 A0A0C9QBD0 Q8MQP1 A0A1W4UM00 A0A0K8W0Z1 Q9W1Q5 A0A182YQ03 A0A0K8W5U7 B4P9L4 B4I8L9 A0A0J9RJQ7 A0A1B6HFK3 A0A182K8Y7 B3NPD7 B4NMN4 A0A182R082 A0A1S4F3N2 A0A182PGE7 A0A182HMG2 Q7Q7F2 A0A182KW16 Q0IG60 A0A182R2K5 B0VZ36 A0A182XD31 A0A1Q3F5H2 A0A182WAK8 A0A182LU14 A0A182UUC2 E0VQU2 A0A182U189 A0A182GLL5 A0A182GE88 A0A1W4XLG9 A0A2M4CP47
A0A2J7Q7Z6 A0A2J7Q807 A0A2J7Q7Z1 A0A0L7QXL7 A0A2A3EAI8 A0A088A308 A0A0M9A7H6 E2AZD0 A0A310SGJ3 A0A026WSR1 A0A3L8D8I8 F4WG80 A0A158NT53 A0A154P5F4 A0A195EFH9 A0A151WJE6 A0A195EZR6 A0A195B6B8 A0A151IAK8 A0A0K8VSR4 A0A0K8W2L2 A0A0K8USI9 A0A0K8V3W1 A0A0A1WNI7 A0A0K8TWV5 E2BVS2 A0A1Y1MEH0 A0A0J7KZ56 A0A1B0FPN6 A0A1A9W9M3 A0A1A9VST1 A0A1B0AAY7 A0A1I8NY12 A0A0L0CBA8 A0A1B0BS51 A0A1A9YGH7 A0A1I8N8B6 T1PHB9 A0A034VIG4 A0A0P4VWJ2 D6WTF9 A0A0T6BGP8 B4LMB8 B4J7D8 A0A0Q9X988 B4KSF1 B4GHN8 A0A0M3QV37 Q28ZJ5 B3MF07 A0A3B0JMI2 A0A2P2I2Q4 A0A0C9QBD0 Q8MQP1 A0A1W4UM00 A0A0K8W0Z1 Q9W1Q5 A0A182YQ03 A0A0K8W5U7 B4P9L4 B4I8L9 A0A0J9RJQ7 A0A1B6HFK3 A0A182K8Y7 B3NPD7 B4NMN4 A0A182R082 A0A1S4F3N2 A0A182PGE7 A0A182HMG2 Q7Q7F2 A0A182KW16 Q0IG60 A0A182R2K5 B0VZ36 A0A182XD31 A0A1Q3F5H2 A0A182WAK8 A0A182LU14 A0A182UUC2 E0VQU2 A0A182U189 A0A182GLL5 A0A182GE88 A0A1W4XLG9 A0A2M4CP47
Ontologies
GO
PANTHER
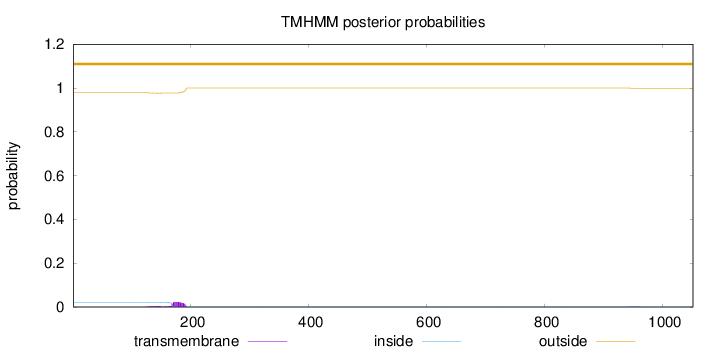
Topology
Length:
1051
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.59565
Exp number, first 60 AAs:
0.00222
Total prob of N-in:
0.02061
outside
1 - 1051
Population Genetic Test Statistics
Pi
222.159824
Theta
182.335866
Tajima's D
0.884532
CLR
0.184081
CSRT
0.625418729063547
Interpretation
Uncertain
