Gene
KWMTBOMO11193 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012660
Annotation
PREDICTED:_3-ketoacyl-CoA_thiolase?_mitochondrial-like_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.263
Sequence
CDS
ATGGCCGTCGCTGTCAACAAAGGCATCTTCATAGTCGCGGCGAAGAGGACCCCGTTCGGAAAAATGGGGGGTCTGTTGAAAGACATTCATCCATCCGACTTACTAGCAGTCGCCGCTAAGGAGTCTTTCAAGGCCGGAAATGTTGCTCCTGCTTTGGTCGACACTGTCAACATTGGACAGGTTTATTCGCTAAGCGGAACATCAGACGGTGGTTTATCACCCCGTCACGCAGCCCTGAAATCAGGCGTGCCACAAGAAAAACCTGCACTGGGCGTCAACAGATTGTGTGGATCAGGATTCCAAGCTGTCATTAACAGTGCCCAAGATATTTTAACAGGTACGGCTCAGATTTCCTTAGCAGGTGGCACGGAGAACATGTCAGCAGTTCCGTTTGTCGTCAGGAACACCCGTTTCGGAGTACCTCTGGGCGTGAATATTCCCTTCGAAGATTCCCTCTCAGCGGGATCTCTGGACACATACTGCAACTACACTATGCCTCAAACTGCTGAAAATTTGGCTGAGAAATACGAACTCAAAAGGGGAGAAGTTGATCAGTTTTCGCTTCAATCCCAGCAGAGATGGAAAACAGCGCACGACAGCGGCGTCTTCAAGGCTGAGATGGCCCCGGTTACAGTCAAAGTGAAAAGGCAAGAAGTACTGGTGGACAGAGACGAGCATCCTAGACCGGAGACCAACACCGAATCGCTAAGTAAATTACCAGCGTTGTTCAGGAAGGGAGGAGTAGTGACGGCTGGAAATTCCTCGGGCGTAAATGATGGTGCCGGTACTGTGATTCTCGCCAGCGAAGAAGCTGTCAAGCAACACGGTCTGACTCCACTGGTTCGTCTCCTCGGCTGGTCGTTTGTTGGTGTGGACCCCAGCATTATGGGTATAGGACCTGTTCCCGCCATCAACAATCTACTATCCGCCACTGGGCTTAAACTTGGCGATATTGACTTGGTAGAGATTAATGAAGCGTTCGCCGCTCAGACTCTAGCTTGTATTAAAGAGCTAAACTTAGACGAAAGTAAATTAAATGTGAACGGAGGGGCCATCGCTATGGGACATCCTGTTGGCGCGTCTGGGGCCAGGATCACAGCTCATTTAGCTCATGAGCTCAGACGTCGTGGTTTGAAGAGAGGTATCGGCTCAGCATGCATTGGAGGAGGGCAAGGAATCGCTTTGCTACTCGAAACTATATAA
Protein
MAVAVNKGIFIVAAKRTPFGKMGGLLKDIHPSDLLAVAAKESFKAGNVAPALVDTVNIGQVYSLSGTSDGGLSPRHAALKSGVPQEKPALGVNRLCGSGFQAVINSAQDILTGTAQISLAGGTENMSAVPFVVRNTRFGVPLGVNIPFEDSLSAGSLDTYCNYTMPQTAENLAEKYELKRGEVDQFSLQSQQRWKTAHDSGVFKAEMAPVTVKVKRQEVLVDRDEHPRPETNTESLSKLPALFRKGGVVTAGNSSGVNDGAGTVILASEEAVKQHGLTPLVRLLGWSFVGVDPSIMGIGPVPAINNLLSATGLKLGDIDLVEINEAFAAQTLACIKELNLDESKLNVNGGAIAMGHPVGASGARITAHLAHELRRRGLKRGIGSACIGGGQGIALLLETI
Summary
Similarity
Belongs to the thiolase-like superfamily. Thiolase family.
Uniprot
H9JSZ8
A0A0P0EA86
A0A2H1W0C5
A0A1V0M8A7
A0A3S2LBN8
A0A2W1BMK7
+ More
A0A291P148 A0A291P0Z9 M4M672 A0A2A4JWP2 M4M667 A0A0L7LDR5 A0A088MCZ0 A0A212ELV4 A0A1Q3G5I8 A0A194QVJ5 A0A2W1BK72 A0A1Q3G5J3 A0A2A4JVH8 A0A1L8D6E7 B7XEI5 M4M6W2 A0A088M9J5 A0A2H1W0G0 A0A2A4JUV2 M4M7X3 A0A291P0X8 A0A291P134 A0A0P0E5G4 A0A1V0M8A3 A0A2H1W083 I4DP15 A0A224XDT7 C6L8Q2 I4DN30 A0A154PTM8 A0A3L8DQK3 M4M648 A0A194QF73 A0A0P4VLL1 A0A1B6C846 R4FQS6 A0A2J7R5D0 A0A1S3DBF4 A0A1W4WLY9 A0A1S3DA08 A0A088AHJ0 K7IV43 D6WSI8 E2ARH3 U5EL67 A0A232FGB8 A0A182N7D7 A0A2S2R8V6 A0A182QTK1 A0A151K395 A0A182Y864 A0A0A9X037 A0A336MAM4 A0A182WB89 A0A151IDY0 A0A2M3ZGQ5 Q1HQG3 A0A2A3EA37 A0A182FFL4 A0A0K8TLC7 A0A2M3Z3U7 J9K576 A0A2M3Z4A9 T1DSQ4 A0A2M4BPZ5 A0A2M4AAT1 A0A1L8DZ48 A0A2M4AAZ6 A0A1L8DZ65 A0A182VD17 A0A182U0X2 A0A182X297 A0A182L3Y7 Q7QIF7 A0A182I6F8 B4MZD5 A0A2M4BQ82 A0A158NQB2 A0A2M4BPY8 A0A195FU57 F4WA98 W5J4S4 A0A182MLQ9 A0A2H8TYJ7 A0A182K4B8 B0W5M7 A0A182RUF2 A0A182IJR5 A0A1Q3FEW7 A0A067RE26 A0A182G6S7 A0A0L0CH53 A0A023ERH3 A0A0A1XGB7
A0A291P148 A0A291P0Z9 M4M672 A0A2A4JWP2 M4M667 A0A0L7LDR5 A0A088MCZ0 A0A212ELV4 A0A1Q3G5I8 A0A194QVJ5 A0A2W1BK72 A0A1Q3G5J3 A0A2A4JVH8 A0A1L8D6E7 B7XEI5 M4M6W2 A0A088M9J5 A0A2H1W0G0 A0A2A4JUV2 M4M7X3 A0A291P0X8 A0A291P134 A0A0P0E5G4 A0A1V0M8A3 A0A2H1W083 I4DP15 A0A224XDT7 C6L8Q2 I4DN30 A0A154PTM8 A0A3L8DQK3 M4M648 A0A194QF73 A0A0P4VLL1 A0A1B6C846 R4FQS6 A0A2J7R5D0 A0A1S3DBF4 A0A1W4WLY9 A0A1S3DA08 A0A088AHJ0 K7IV43 D6WSI8 E2ARH3 U5EL67 A0A232FGB8 A0A182N7D7 A0A2S2R8V6 A0A182QTK1 A0A151K395 A0A182Y864 A0A0A9X037 A0A336MAM4 A0A182WB89 A0A151IDY0 A0A2M3ZGQ5 Q1HQG3 A0A2A3EA37 A0A182FFL4 A0A0K8TLC7 A0A2M3Z3U7 J9K576 A0A2M3Z4A9 T1DSQ4 A0A2M4BPZ5 A0A2M4AAT1 A0A1L8DZ48 A0A2M4AAZ6 A0A1L8DZ65 A0A182VD17 A0A182U0X2 A0A182X297 A0A182L3Y7 Q7QIF7 A0A182I6F8 B4MZD5 A0A2M4BQ82 A0A158NQB2 A0A2M4BPY8 A0A195FU57 F4WA98 W5J4S4 A0A182MLQ9 A0A2H8TYJ7 A0A182K4B8 B0W5M7 A0A182RUF2 A0A182IJR5 A0A1Q3FEW7 A0A067RE26 A0A182G6S7 A0A0L0CH53 A0A023ERH3 A0A0A1XGB7
Pubmed
19121390
26445454
28349297
28756777
28986331
23294019
+ More
26227816 26385554 22118469 26354079 22651552 30249741 27129103 20075255 18362917 19820115 20798317 28648823 25244985 25401762 26823975 17204158 26369729 20966253 12364791 14747013 17210077 17994087 21347285 21719571 20920257 23761445 24845553 26483478 26108605 24945155 25830018
26227816 26385554 22118469 26354079 22651552 30249741 27129103 20075255 18362917 19820115 20798317 28648823 25244985 25401762 26823975 17204158 26369729 20966253 12364791 14747013 17210077 17994087 21347285 21719571 20920257 23761445 24845553 26483478 26108605 24945155 25830018
EMBL
BABH01038945
KT261728
ALJ30268.1
ODYU01005556
SOQ46518.1
KU755510
+ More
ARD71220.1 RSAL01000270 RVE43181.1 KZ150068 PZC74106.1 MF706185 ATJ44612.1 MF687661 ATJ44587.1 JX847000 AGG55017.1 NWSH01000536 PCG75820.1 JX846985 AGG55002.1 JTDY01001527 KOB73648.1 KJ579207 AIN34683.1 AGBW02013975 OWR42472.1 GEYN01000173 JAV44816.1 KQ461073 KPJ09492.1 PZC74105.1 GEYN01000177 JAV44812.1 PCG75819.1 GEYN01000164 JAV01965.1 AB466326 BAH03386.1 JX846982 AGG54999.1 KJ579213 AIN34689.1 SOQ46517.1 PCG75817.1 JX846998 AGG55015.1 MF687641 ATJ44567.1 MF706169 ATJ44596.1 PZC74103.1 KT261708 ALJ30248.1 KU755502 ARD71212.1 SOQ46515.1 AK403244 BAM19655.1 GFTR01005790 JAW10636.1 AB516425 BAH96561.1 AK402698 BAM19320.1 KQ435119 KZC14708.1 QOIP01000006 RLU22088.1 JX846984 AGG55001.1 KQ459053 KPJ04132.1 GDKW01001282 JAI55313.1 GEDC01027694 JAS09604.1 GAHY01000244 JAA77266.1 NEVH01006994 PNF36030.1 AAZX01001202 KQ971354 EFA05913.1 GL442063 EFN63982.1 GANO01001545 JAB58326.1 NNAY01000245 OXU29742.1 GGMS01016917 MBY86120.1 AXCN02000568 LKEY01022044 KYN50547.1 GBHO01031506 GBRD01004592 GDHC01001442 JAG12098.1 JAG61229.1 JAQ17187.1 UFQT01000822 SSX27392.1 KQ977901 KYM98885.1 GGFM01006985 MBW27736.1 DQ440481 ABF18514.1 KZ288310 PBC28595.1 GDAI01002637 JAI14966.1 GGFM01002443 MBW23194.1 ABLF02027851 GGFM01002582 MBW23333.1 GAMD01001374 JAB00217.1 GGFJ01006014 MBW55155.1 GGFK01004566 MBW37887.1 GFDF01002378 JAV11706.1 GGFK01004561 MBW37882.1 GFDF01002377 JAV11707.1 AAAB01008807 EAA04232.2 APCN01003635 CH963913 EDW77408.1 GGFJ01006105 MBW55246.1 ADTU01023023 ADTU01023024 GGFJ01006015 MBW55156.1 KQ981272 KYN43827.1 GL888045 EGI68889.1 ADMH02002207 ETN57809.1 AXCM01006679 GFXV01007549 MBW19354.1 DS231843 EDS35510.1 GFDL01008949 JAV26096.1 KK852527 KDR22017.1 JXUM01045461 KQ561439 KXJ78588.1 JRES01000384 KNC31743.1 GAPW01001820 JAC11778.1 GBXI01004280 JAD10012.1
ARD71220.1 RSAL01000270 RVE43181.1 KZ150068 PZC74106.1 MF706185 ATJ44612.1 MF687661 ATJ44587.1 JX847000 AGG55017.1 NWSH01000536 PCG75820.1 JX846985 AGG55002.1 JTDY01001527 KOB73648.1 KJ579207 AIN34683.1 AGBW02013975 OWR42472.1 GEYN01000173 JAV44816.1 KQ461073 KPJ09492.1 PZC74105.1 GEYN01000177 JAV44812.1 PCG75819.1 GEYN01000164 JAV01965.1 AB466326 BAH03386.1 JX846982 AGG54999.1 KJ579213 AIN34689.1 SOQ46517.1 PCG75817.1 JX846998 AGG55015.1 MF687641 ATJ44567.1 MF706169 ATJ44596.1 PZC74103.1 KT261708 ALJ30248.1 KU755502 ARD71212.1 SOQ46515.1 AK403244 BAM19655.1 GFTR01005790 JAW10636.1 AB516425 BAH96561.1 AK402698 BAM19320.1 KQ435119 KZC14708.1 QOIP01000006 RLU22088.1 JX846984 AGG55001.1 KQ459053 KPJ04132.1 GDKW01001282 JAI55313.1 GEDC01027694 JAS09604.1 GAHY01000244 JAA77266.1 NEVH01006994 PNF36030.1 AAZX01001202 KQ971354 EFA05913.1 GL442063 EFN63982.1 GANO01001545 JAB58326.1 NNAY01000245 OXU29742.1 GGMS01016917 MBY86120.1 AXCN02000568 LKEY01022044 KYN50547.1 GBHO01031506 GBRD01004592 GDHC01001442 JAG12098.1 JAG61229.1 JAQ17187.1 UFQT01000822 SSX27392.1 KQ977901 KYM98885.1 GGFM01006985 MBW27736.1 DQ440481 ABF18514.1 KZ288310 PBC28595.1 GDAI01002637 JAI14966.1 GGFM01002443 MBW23194.1 ABLF02027851 GGFM01002582 MBW23333.1 GAMD01001374 JAB00217.1 GGFJ01006014 MBW55155.1 GGFK01004566 MBW37887.1 GFDF01002378 JAV11706.1 GGFK01004561 MBW37882.1 GFDF01002377 JAV11707.1 AAAB01008807 EAA04232.2 APCN01003635 CH963913 EDW77408.1 GGFJ01006105 MBW55246.1 ADTU01023023 ADTU01023024 GGFJ01006015 MBW55156.1 KQ981272 KYN43827.1 GL888045 EGI68889.1 ADMH02002207 ETN57809.1 AXCM01006679 GFXV01007549 MBW19354.1 DS231843 EDS35510.1 GFDL01008949 JAV26096.1 KK852527 KDR22017.1 JXUM01045461 KQ561439 KXJ78588.1 JRES01000384 KNC31743.1 GAPW01001820 JAC11778.1 GBXI01004280 JAD10012.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000037510
UP000007151
UP000053240
+ More
UP000076502 UP000279307 UP000053268 UP000235965 UP000079169 UP000192223 UP000005203 UP000002358 UP000007266 UP000000311 UP000215335 UP000075884 UP000075886 UP000078492 UP000076408 UP000075920 UP000078542 UP000242457 UP000069272 UP000007819 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000007798 UP000005205 UP000078541 UP000007755 UP000000673 UP000075883 UP000075881 UP000002320 UP000075900 UP000075880 UP000027135 UP000069940 UP000249989 UP000037069
UP000076502 UP000279307 UP000053268 UP000235965 UP000079169 UP000192223 UP000005203 UP000002358 UP000007266 UP000000311 UP000215335 UP000075884 UP000075886 UP000078492 UP000076408 UP000075920 UP000078542 UP000242457 UP000069272 UP000007819 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000007798 UP000005205 UP000078541 UP000007755 UP000000673 UP000075883 UP000075881 UP000002320 UP000075900 UP000075880 UP000027135 UP000069940 UP000249989 UP000037069
PRIDE
Interpro
SUPFAM
SSF53901
SSF53901
Gene 3D
CDD
ProteinModelPortal
H9JSZ8
A0A0P0EA86
A0A2H1W0C5
A0A1V0M8A7
A0A3S2LBN8
A0A2W1BMK7
+ More
A0A291P148 A0A291P0Z9 M4M672 A0A2A4JWP2 M4M667 A0A0L7LDR5 A0A088MCZ0 A0A212ELV4 A0A1Q3G5I8 A0A194QVJ5 A0A2W1BK72 A0A1Q3G5J3 A0A2A4JVH8 A0A1L8D6E7 B7XEI5 M4M6W2 A0A088M9J5 A0A2H1W0G0 A0A2A4JUV2 M4M7X3 A0A291P0X8 A0A291P134 A0A0P0E5G4 A0A1V0M8A3 A0A2H1W083 I4DP15 A0A224XDT7 C6L8Q2 I4DN30 A0A154PTM8 A0A3L8DQK3 M4M648 A0A194QF73 A0A0P4VLL1 A0A1B6C846 R4FQS6 A0A2J7R5D0 A0A1S3DBF4 A0A1W4WLY9 A0A1S3DA08 A0A088AHJ0 K7IV43 D6WSI8 E2ARH3 U5EL67 A0A232FGB8 A0A182N7D7 A0A2S2R8V6 A0A182QTK1 A0A151K395 A0A182Y864 A0A0A9X037 A0A336MAM4 A0A182WB89 A0A151IDY0 A0A2M3ZGQ5 Q1HQG3 A0A2A3EA37 A0A182FFL4 A0A0K8TLC7 A0A2M3Z3U7 J9K576 A0A2M3Z4A9 T1DSQ4 A0A2M4BPZ5 A0A2M4AAT1 A0A1L8DZ48 A0A2M4AAZ6 A0A1L8DZ65 A0A182VD17 A0A182U0X2 A0A182X297 A0A182L3Y7 Q7QIF7 A0A182I6F8 B4MZD5 A0A2M4BQ82 A0A158NQB2 A0A2M4BPY8 A0A195FU57 F4WA98 W5J4S4 A0A182MLQ9 A0A2H8TYJ7 A0A182K4B8 B0W5M7 A0A182RUF2 A0A182IJR5 A0A1Q3FEW7 A0A067RE26 A0A182G6S7 A0A0L0CH53 A0A023ERH3 A0A0A1XGB7
A0A291P148 A0A291P0Z9 M4M672 A0A2A4JWP2 M4M667 A0A0L7LDR5 A0A088MCZ0 A0A212ELV4 A0A1Q3G5I8 A0A194QVJ5 A0A2W1BK72 A0A1Q3G5J3 A0A2A4JVH8 A0A1L8D6E7 B7XEI5 M4M6W2 A0A088M9J5 A0A2H1W0G0 A0A2A4JUV2 M4M7X3 A0A291P0X8 A0A291P134 A0A0P0E5G4 A0A1V0M8A3 A0A2H1W083 I4DP15 A0A224XDT7 C6L8Q2 I4DN30 A0A154PTM8 A0A3L8DQK3 M4M648 A0A194QF73 A0A0P4VLL1 A0A1B6C846 R4FQS6 A0A2J7R5D0 A0A1S3DBF4 A0A1W4WLY9 A0A1S3DA08 A0A088AHJ0 K7IV43 D6WSI8 E2ARH3 U5EL67 A0A232FGB8 A0A182N7D7 A0A2S2R8V6 A0A182QTK1 A0A151K395 A0A182Y864 A0A0A9X037 A0A336MAM4 A0A182WB89 A0A151IDY0 A0A2M3ZGQ5 Q1HQG3 A0A2A3EA37 A0A182FFL4 A0A0K8TLC7 A0A2M3Z3U7 J9K576 A0A2M3Z4A9 T1DSQ4 A0A2M4BPZ5 A0A2M4AAT1 A0A1L8DZ48 A0A2M4AAZ6 A0A1L8DZ65 A0A182VD17 A0A182U0X2 A0A182X297 A0A182L3Y7 Q7QIF7 A0A182I6F8 B4MZD5 A0A2M4BQ82 A0A158NQB2 A0A2M4BPY8 A0A195FU57 F4WA98 W5J4S4 A0A182MLQ9 A0A2H8TYJ7 A0A182K4B8 B0W5M7 A0A182RUF2 A0A182IJR5 A0A1Q3FEW7 A0A067RE26 A0A182G6S7 A0A0L0CH53 A0A023ERH3 A0A0A1XGB7
PDB
4C2K
E-value=7.67606e-116,
Score=1067
Ontologies
PATHWAY
00062
Fatty acid elongation - Bombyx mori (domestic silkworm)
00071 Fatty acid degradation - Bombyx mori (domestic silkworm)
00280 Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01212 Fatty acid metabolism - Bombyx mori (domestic silkworm)
00071 Fatty acid degradation - Bombyx mori (domestic silkworm)
00280 Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01212 Fatty acid metabolism - Bombyx mori (domestic silkworm)
GO
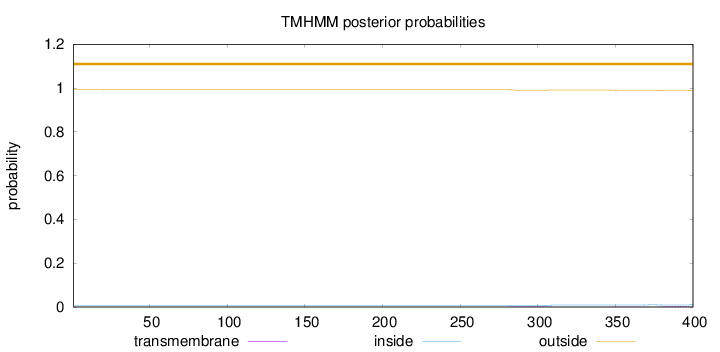
Topology
Length:
400
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.15636
Exp number, first 60 AAs:
0.00284
Total prob of N-in:
0.00819
outside
1 - 400
Population Genetic Test Statistics
Pi
179.264384
Theta
156.175129
Tajima's D
0.464844
CLR
0.158243
CSRT
0.503424828758562
Interpretation
Uncertain
