Gene
KWMTBOMO11190
Pre Gene Modal
BGIBMGA012658
Annotation
PREDICTED:_CCR4-NOT_transcription_complex_subunit_11_isoform_X1_[Papilio_machaon]
Location in the cell
Cytoplasmic Reliability : 1.743 Nuclear Reliability : 1.044
Sequence
CDS
ATGTCTCAACATCTGATTAATGAAAACAGTAAATACATACTCGATTTATTTTCCGAACAGACAGTCGATTCGCAGGGCTTAGATTCAATATGTGTTCAAGTTCAAAAAAAGTTCCCGAAGACCGATCATTTTAATCTAAGTCTGCTCCTGTCATCGCTGATTACGGGCGGAGATCTTTCACTCCCTGGGCAGCGTGTTGTTGCACTGGCTCTTCTATATGACTTCTACAAGGGAGAAAACCCATTCAGCCCATTGTTCTTGCATCTCCTAGAAGGGAAGCTGGGTCTTCTGCCTCTAGCTCCACAGGAGAGACTGTTTATAGGACAGCTGCATGGATTCTTGCCTGTAAACATTAAAGATGTTATGAAAAAATCAGCAAAACAAGTAATGATGACAGAAGTATCTCCTAAGGACCTGGAGTTTGATTACTCACCGCTGCAGGCGATTGTGGCCGAACGTTTGACTGACACCAGTGCTGTGGCCCGGGCCACTTGTTCTGCATTGATACCGCTTAGTGATGGGAGTCCACCGAACCGCCTTGCCTTGAAAGAACTGCTAGAAGCCTTGATGAGTAATGAGTATCTCCCGCTGCACCGAACGCTATCGGCCGCCGGACCCATCCCACCGCCCACGTTCCTCATCGACACCAATGATGTGCCTTTTGCTGACGAGGGCGTTTGGAAGAACTTAATAAATCGCGGCACGTATATACCGATGTATGACTCCGACTACGACGGACTGATCGGGCTCCGACCTGAGAAGATCGATAGAAGATCGCAGCCTAACCAACAGCACAAAGTCAAGGAGGAAAAAAAAGAGAAACAAGAAGAGAAGAAGGAAGATAAAGATCCTGTGTCATTAGTGAGTGAAGCTAAAGATTTGACCCAGATCGCGCTCAGGGGAGCCCTCAGTGTACAGCAACAGCAAAGACTACTGGCACTGCTGGATCAGGAACCTAACATTGTCTACGAAATAGGTGTTACACCTAACCAGCTACCGGACCTAGTCGAGAACAATCCAATGGTCGCCATATCGGTGCTTCTCAAACTGATCCACTCTCAGCACATCACGGAGTACTTCAGTGTCCTCGTCAACATGGAAATGTCGCTGCACTCCATGGAGGTGGTCAACAGGTTAACGACCTCAGTGGATTTGCCGGTGGAGTTCGTTCACCTCTACATAAGCAATTGTATTTCGACGTGTGAGACAATACGGGACAGATACATGCAAAACAGGCTCGTACGATTGGTGTGTGTCTTCCTGCAGTCGCTGATAAGGAACAAGATCATCAATGTGAAGGAGCTATTCATAGAAGTGGAAGCGTTCTGCGTTGAGTTCAGCCGGATCCGAGAGGCCGCTGCGCTGTTTCGGCTCCTGAAGCAGCTGGACTCCGGCGAGGCTGCCCACAAGGACCCCAAGGATAACTAG
Protein
MSQHLINENSKYILDLFSEQTVDSQGLDSICVQVQKKFPKTDHFNLSLLLSSLITGGDLSLPGQRVVALALLYDFYKGENPFSPLFLHLLEGKLGLLPLAPQERLFIGQLHGFLPVNIKDVMKKSAKQVMMTEVSPKDLEFDYSPLQAIVAERLTDTSAVARATCSALIPLSDGSPPNRLALKELLEALMSNEYLPLHRTLSAAGPIPPPTFLIDTNDVPFADEGVWKNLINRGTYIPMYDSDYDGLIGLRPEKIDRRSQPNQQHKVKEEKKEKQEEKKEDKDPVSLVSEAKDLTQIALRGALSVQQQQRLLALLDQEPNIVYEIGVTPNQLPDLVENNPMVAISVLLKLIHSQHITEYFSVLVNMEMSLHSMEVVNRLTTSVDLPVEFVHLYISNCISTCETIRDRYMQNRLVRLVCVFLQSLIRNKIINVKELFIEVEAFCVEFSRIREAAALFRLLKQLDSGEAAHKDPKDN
Summary
Uniprot
H9JSZ6
A0A2H1WQY9
A0A194R0X6
A0A194QFD8
A0A2A4JW04
A0A2W1BG51
+ More
A0A212ELW1 A0A023F3X5 A0A1B6DGH3 A0A069DU08 A0A1B6FCN2 A0A1W4XMW0 A0A224XCB6 D6WTS3 A0A2R7W4L2 A0A0A9XZQ0 A0A0A9XWP2 A0A146M0U8 A0A067QJL2 U4U497 J3JW98 A0A1Y1LS00 A0A2P8XVN3 A0A2A3E2B0 A0A087ZXS5 A0A2J7RGU6 E2BU53 A0A1W2WI25 A0A0P5ZBF4 A0A151WNX2 V5GSH0 A0A0N7ZDX7 A0A224YPR4 A0A131Z5D1 A0A232F273 A0A0P5LNA1 A0A0P5YS75 K7J4T2 V5I0U1 A0A0P6AM58 A0A3M6UDZ7 A0A0P5RRQ0 A0A0N7ZQG8 E2AW92 A0A2R5LFQ1 A0A1E1XAN1 A0A0J7K8N7 A7RPS5 A0A0P5Z5D3 A0A151NXT5 A0A2K6G1Z7 F6RII5 A0A091I7G0 A0A2K6G1Y1 F6YYW3 F6Y9Y2 A0A091FH32 A0A3Q1IU27 A0A1A8GWG7 A0A2K5JJV6 F1STG2 A0A3Q3M163 A0A1L8HGQ5 D2HBE4 A0A3M0K0Z5 A0A2Y9D8A7 A0A3Q3D2L6 A0A1A8S278 A0A1A8QJI8 A0A091FTY0 A0A2I0UCR6 A0A226MFH1 A0A226P6N9 A0A1V4J3G9 A0A093Q3T9 A0A2I0MSY2 G1MAR3 A0A093G9Q8 A0A2I3GWI2 A0A2K5QUD7 G1RWI2 A0A2K6JQQ9 H9GCZ2 A0A2K5QUE6 A0A1A7Y2E4 A0A2K5DI19 A0A2K5DI45 A0A2K6U6N0 F6U2U4 A0A2K6E5A1 A0A2K5VMN5 A0A2I3N1D3 U3D6A6 U3JMG4 A0A3L8STH9 A0A218UWT8 A0A3Q7W118
A0A212ELW1 A0A023F3X5 A0A1B6DGH3 A0A069DU08 A0A1B6FCN2 A0A1W4XMW0 A0A224XCB6 D6WTS3 A0A2R7W4L2 A0A0A9XZQ0 A0A0A9XWP2 A0A146M0U8 A0A067QJL2 U4U497 J3JW98 A0A1Y1LS00 A0A2P8XVN3 A0A2A3E2B0 A0A087ZXS5 A0A2J7RGU6 E2BU53 A0A1W2WI25 A0A0P5ZBF4 A0A151WNX2 V5GSH0 A0A0N7ZDX7 A0A224YPR4 A0A131Z5D1 A0A232F273 A0A0P5LNA1 A0A0P5YS75 K7J4T2 V5I0U1 A0A0P6AM58 A0A3M6UDZ7 A0A0P5RRQ0 A0A0N7ZQG8 E2AW92 A0A2R5LFQ1 A0A1E1XAN1 A0A0J7K8N7 A7RPS5 A0A0P5Z5D3 A0A151NXT5 A0A2K6G1Z7 F6RII5 A0A091I7G0 A0A2K6G1Y1 F6YYW3 F6Y9Y2 A0A091FH32 A0A3Q1IU27 A0A1A8GWG7 A0A2K5JJV6 F1STG2 A0A3Q3M163 A0A1L8HGQ5 D2HBE4 A0A3M0K0Z5 A0A2Y9D8A7 A0A3Q3D2L6 A0A1A8S278 A0A1A8QJI8 A0A091FTY0 A0A2I0UCR6 A0A226MFH1 A0A226P6N9 A0A1V4J3G9 A0A093Q3T9 A0A2I0MSY2 G1MAR3 A0A093G9Q8 A0A2I3GWI2 A0A2K5QUD7 G1RWI2 A0A2K6JQQ9 H9GCZ2 A0A2K5QUE6 A0A1A7Y2E4 A0A2K5DI19 A0A2K5DI45 A0A2K6U6N0 F6U2U4 A0A2K6E5A1 A0A2K5VMN5 A0A2I3N1D3 U3D6A6 U3JMG4 A0A3L8STH9 A0A218UWT8 A0A3Q7W118
Pubmed
19121390
26354079
28756777
22118469
25474469
26334808
+ More
18362917 19820115 25401762 26823975 24845553 23537049 22516182 28004739 29403074 20798317 28797301 26830274 28648823 20075255 25765539 30382153 28503490 17615350 22293439 18464734 12481130 15114417 20431018 30723633 27762356 20010809 24621616 23371554 21881562 19892987 25243066 30282656
18362917 19820115 25401762 26823975 24845553 23537049 22516182 28004739 29403074 20798317 28797301 26830274 28648823 20075255 25765539 30382153 28503490 17615350 22293439 18464734 12481130 15114417 20431018 30723633 27762356 20010809 24621616 23371554 21881562 19892987 25243066 30282656
EMBL
BABH01038942
BABH01038943
ODYU01010351
SOQ55396.1
KQ461073
KPJ09496.1
+ More
KQ459053 KPJ04129.1 NWSH01000536 PCG75814.1 KZ150068 PZC74099.1 AGBW02013975 OWR42475.1 GBBI01003063 JAC15649.1 GEDC01025455 GEDC01023514 GEDC01012578 JAS11843.1 JAS13784.1 JAS24720.1 GBGD01001504 JAC87385.1 GECZ01022056 JAS47713.1 GFTR01006399 JAW10027.1 KQ971352 EFA06281.1 KK854300 PTY14368.1 GBHO01019271 JAG24333.1 GBHO01019270 JAG24334.1 GDHC01006232 JAQ12397.1 KK853284 KDR08992.1 KB632095 ERL88729.1 APGK01025938 BT127516 KB740605 AEE62478.1 ENN80068.1 GEZM01049935 JAV75751.1 PYGN01001278 PSN36077.1 KZ288430 PBC25830.1 NEVH01003751 PNF40061.1 GL450581 EFN80770.1 GDIP01046020 LRGB01000725 JAM57695.1 KZS16375.1 KQ982892 KYQ49602.1 GALX01005278 JAB63188.1 GDIP01254921 JAI68480.1 GFPF01004684 MAA15830.1 GEDV01002379 JAP86178.1 NNAY01001281 OXU24507.1 GDIQ01167418 JAK84307.1 GDIP01054315 JAM49400.1 AAZX01000449 GANP01003615 JAB80853.1 GDIP01032986 JAM70729.1 RCHS01001707 RMX51900.1 GDIQ01102796 JAL48930.1 GDIP01222593 JAJ00809.1 GL443262 EFN62314.1 GGLE01004162 MBY08288.1 GFAC01002916 JAT96272.1 LBMM01011672 KMQ86707.1 DS469526 EDO46626.1 GDIP01048956 JAM54759.1 AKHW03001628 KYO41409.1 KL218306 KFP04172.1 EAAA01001446 AAMC01034846 KK718990 KFO60775.1 HAEB01016745 HAEC01007274 SBQ75412.1 AEMK02000020 DQIR01113895 DQIR01193510 HDA69371.1 CM004468 OCT95256.1 GL192657 EFB13197.1 QRBI01000120 RMC06648.1 HAEI01005885 HAEH01021265 SBS12062.1 HAEF01002966 HAEG01012936 SBR93627.1 KL447476 KFO73980.1 KZ505871 PKU43783.1 MCFN01000999 OXB54042.1 AWGT02000157 OXB75157.1 LSYS01009367 OPJ66756.1 KL671095 KFW81115.1 AKCR02000003 PKK32790.1 ACTA01122438 ACTA01130438 ACTA01138438 KL215166 KFV63607.1 ADFV01084183 AAWZ02029971 HADW01007457 HADX01002244 SBP24476.1 AQIA01016841 AHZZ02005972 AHZZ02005973 AHZZ02005974 GAMT01004081 GAMS01001307 GAMR01006305 GAMQ01003276 GAMQ01003275 GAMP01008801 JAB07780.1 JAB21829.1 JAB27627.1 JAB38575.1 JAB43954.1 AGTO01000128 QUSF01000007 RLW07705.1 MUZQ01000105 OWK58233.1
KQ459053 KPJ04129.1 NWSH01000536 PCG75814.1 KZ150068 PZC74099.1 AGBW02013975 OWR42475.1 GBBI01003063 JAC15649.1 GEDC01025455 GEDC01023514 GEDC01012578 JAS11843.1 JAS13784.1 JAS24720.1 GBGD01001504 JAC87385.1 GECZ01022056 JAS47713.1 GFTR01006399 JAW10027.1 KQ971352 EFA06281.1 KK854300 PTY14368.1 GBHO01019271 JAG24333.1 GBHO01019270 JAG24334.1 GDHC01006232 JAQ12397.1 KK853284 KDR08992.1 KB632095 ERL88729.1 APGK01025938 BT127516 KB740605 AEE62478.1 ENN80068.1 GEZM01049935 JAV75751.1 PYGN01001278 PSN36077.1 KZ288430 PBC25830.1 NEVH01003751 PNF40061.1 GL450581 EFN80770.1 GDIP01046020 LRGB01000725 JAM57695.1 KZS16375.1 KQ982892 KYQ49602.1 GALX01005278 JAB63188.1 GDIP01254921 JAI68480.1 GFPF01004684 MAA15830.1 GEDV01002379 JAP86178.1 NNAY01001281 OXU24507.1 GDIQ01167418 JAK84307.1 GDIP01054315 JAM49400.1 AAZX01000449 GANP01003615 JAB80853.1 GDIP01032986 JAM70729.1 RCHS01001707 RMX51900.1 GDIQ01102796 JAL48930.1 GDIP01222593 JAJ00809.1 GL443262 EFN62314.1 GGLE01004162 MBY08288.1 GFAC01002916 JAT96272.1 LBMM01011672 KMQ86707.1 DS469526 EDO46626.1 GDIP01048956 JAM54759.1 AKHW03001628 KYO41409.1 KL218306 KFP04172.1 EAAA01001446 AAMC01034846 KK718990 KFO60775.1 HAEB01016745 HAEC01007274 SBQ75412.1 AEMK02000020 DQIR01113895 DQIR01193510 HDA69371.1 CM004468 OCT95256.1 GL192657 EFB13197.1 QRBI01000120 RMC06648.1 HAEI01005885 HAEH01021265 SBS12062.1 HAEF01002966 HAEG01012936 SBR93627.1 KL447476 KFO73980.1 KZ505871 PKU43783.1 MCFN01000999 OXB54042.1 AWGT02000157 OXB75157.1 LSYS01009367 OPJ66756.1 KL671095 KFW81115.1 AKCR02000003 PKK32790.1 ACTA01122438 ACTA01130438 ACTA01138438 KL215166 KFV63607.1 ADFV01084183 AAWZ02029971 HADW01007457 HADX01002244 SBP24476.1 AQIA01016841 AHZZ02005972 AHZZ02005973 AHZZ02005974 GAMT01004081 GAMS01001307 GAMR01006305 GAMQ01003276 GAMQ01003275 GAMP01008801 JAB07780.1 JAB21829.1 JAB27627.1 JAB38575.1 JAB43954.1 AGTO01000128 QUSF01000007 RLW07705.1 MUZQ01000105 OWK58233.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000192223
+ More
UP000007266 UP000027135 UP000030742 UP000019118 UP000245037 UP000242457 UP000005203 UP000235965 UP000008237 UP000076858 UP000075809 UP000215335 UP000002358 UP000275408 UP000000311 UP000036403 UP000001593 UP000050525 UP000233160 UP000002279 UP000054308 UP000008144 UP000008143 UP000052976 UP000265040 UP000233080 UP000008227 UP000261640 UP000186698 UP000269221 UP000248480 UP000264820 UP000053760 UP000198323 UP000198419 UP000190648 UP000053258 UP000053872 UP000008912 UP000053875 UP000001073 UP000233040 UP000233180 UP000001646 UP000233020 UP000233220 UP000002281 UP000233120 UP000233100 UP000028761 UP000008225 UP000016665 UP000276834 UP000197619 UP000286642
UP000007266 UP000027135 UP000030742 UP000019118 UP000245037 UP000242457 UP000005203 UP000235965 UP000008237 UP000076858 UP000075809 UP000215335 UP000002358 UP000275408 UP000000311 UP000036403 UP000001593 UP000050525 UP000233160 UP000002279 UP000054308 UP000008144 UP000008143 UP000052976 UP000265040 UP000233080 UP000008227 UP000261640 UP000186698 UP000269221 UP000248480 UP000264820 UP000053760 UP000198323 UP000198419 UP000190648 UP000053258 UP000053872 UP000008912 UP000053875 UP000001073 UP000233040 UP000233180 UP000001646 UP000233020 UP000233220 UP000002281 UP000233120 UP000233100 UP000028761 UP000008225 UP000016665 UP000276834 UP000197619 UP000286642
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
H9JSZ6
A0A2H1WQY9
A0A194R0X6
A0A194QFD8
A0A2A4JW04
A0A2W1BG51
+ More
A0A212ELW1 A0A023F3X5 A0A1B6DGH3 A0A069DU08 A0A1B6FCN2 A0A1W4XMW0 A0A224XCB6 D6WTS3 A0A2R7W4L2 A0A0A9XZQ0 A0A0A9XWP2 A0A146M0U8 A0A067QJL2 U4U497 J3JW98 A0A1Y1LS00 A0A2P8XVN3 A0A2A3E2B0 A0A087ZXS5 A0A2J7RGU6 E2BU53 A0A1W2WI25 A0A0P5ZBF4 A0A151WNX2 V5GSH0 A0A0N7ZDX7 A0A224YPR4 A0A131Z5D1 A0A232F273 A0A0P5LNA1 A0A0P5YS75 K7J4T2 V5I0U1 A0A0P6AM58 A0A3M6UDZ7 A0A0P5RRQ0 A0A0N7ZQG8 E2AW92 A0A2R5LFQ1 A0A1E1XAN1 A0A0J7K8N7 A7RPS5 A0A0P5Z5D3 A0A151NXT5 A0A2K6G1Z7 F6RII5 A0A091I7G0 A0A2K6G1Y1 F6YYW3 F6Y9Y2 A0A091FH32 A0A3Q1IU27 A0A1A8GWG7 A0A2K5JJV6 F1STG2 A0A3Q3M163 A0A1L8HGQ5 D2HBE4 A0A3M0K0Z5 A0A2Y9D8A7 A0A3Q3D2L6 A0A1A8S278 A0A1A8QJI8 A0A091FTY0 A0A2I0UCR6 A0A226MFH1 A0A226P6N9 A0A1V4J3G9 A0A093Q3T9 A0A2I0MSY2 G1MAR3 A0A093G9Q8 A0A2I3GWI2 A0A2K5QUD7 G1RWI2 A0A2K6JQQ9 H9GCZ2 A0A2K5QUE6 A0A1A7Y2E4 A0A2K5DI19 A0A2K5DI45 A0A2K6U6N0 F6U2U4 A0A2K6E5A1 A0A2K5VMN5 A0A2I3N1D3 U3D6A6 U3JMG4 A0A3L8STH9 A0A218UWT8 A0A3Q7W118
A0A212ELW1 A0A023F3X5 A0A1B6DGH3 A0A069DU08 A0A1B6FCN2 A0A1W4XMW0 A0A224XCB6 D6WTS3 A0A2R7W4L2 A0A0A9XZQ0 A0A0A9XWP2 A0A146M0U8 A0A067QJL2 U4U497 J3JW98 A0A1Y1LS00 A0A2P8XVN3 A0A2A3E2B0 A0A087ZXS5 A0A2J7RGU6 E2BU53 A0A1W2WI25 A0A0P5ZBF4 A0A151WNX2 V5GSH0 A0A0N7ZDX7 A0A224YPR4 A0A131Z5D1 A0A232F273 A0A0P5LNA1 A0A0P5YS75 K7J4T2 V5I0U1 A0A0P6AM58 A0A3M6UDZ7 A0A0P5RRQ0 A0A0N7ZQG8 E2AW92 A0A2R5LFQ1 A0A1E1XAN1 A0A0J7K8N7 A7RPS5 A0A0P5Z5D3 A0A151NXT5 A0A2K6G1Z7 F6RII5 A0A091I7G0 A0A2K6G1Y1 F6YYW3 F6Y9Y2 A0A091FH32 A0A3Q1IU27 A0A1A8GWG7 A0A2K5JJV6 F1STG2 A0A3Q3M163 A0A1L8HGQ5 D2HBE4 A0A3M0K0Z5 A0A2Y9D8A7 A0A3Q3D2L6 A0A1A8S278 A0A1A8QJI8 A0A091FTY0 A0A2I0UCR6 A0A226MFH1 A0A226P6N9 A0A1V4J3G9 A0A093Q3T9 A0A2I0MSY2 G1MAR3 A0A093G9Q8 A0A2I3GWI2 A0A2K5QUD7 G1RWI2 A0A2K6JQQ9 H9GCZ2 A0A2K5QUE6 A0A1A7Y2E4 A0A2K5DI19 A0A2K5DI45 A0A2K6U6N0 F6U2U4 A0A2K6E5A1 A0A2K5VMN5 A0A2I3N1D3 U3D6A6 U3JMG4 A0A3L8STH9 A0A218UWT8 A0A3Q7W118
Ontologies
PANTHER
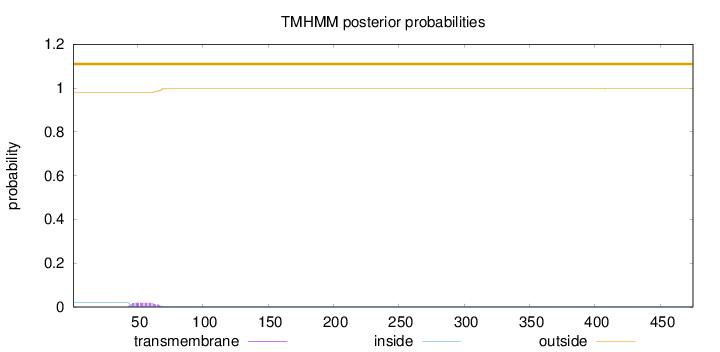
Topology
Length:
475
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.40887
Exp number, first 60 AAs:
0.30085
Total prob of N-in:
0.02199
outside
1 - 475
Population Genetic Test Statistics
Pi
206.024655
Theta
168.809876
Tajima's D
0.940453
CLR
0.06427
CSRT
0.64276786160692
Interpretation
Uncertain
