Gene
KWMTBOMO11161 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008595
Annotation
PREDICTED:_26S_proteasome_non-ATPase_regulatory_subunit_8_[Papilio_machaon]
Full name
26S proteasome non-ATPase regulatory subunit 8
Alternative Name
26S proteasome regulatory subunit RPN12
26S proteasome regulatory subunit S14
26S proteasome regulatory subunit S14
Location in the cell
Cytoplasmic Reliability : 2.182
Sequence
CDS
ATGGCTACTGTAAAAGACGTCGCTACATTATATCAAAATCTGAAGACCGAGTGGGCAAAGAAGCCAAGAAAGCTGGACAAATGTGGAGAGCTCTTAAACAAAATAAAGATTGCTCTTACACAGTTAACGTTTCTTCCAAGTAATAATGTCGCAGCAAACCAAAAAGAGTTGATTTTGGCTAGAGATGTATTGGAAATAGGTGCACAATGGGCCGTCGCCGTGAAAGACGTCAAAGCTTTCGAGAGATACATGTCTCAACTCAAGTGTTATTATTTCGATTACAAAGACCACCTCCCGGAGTCTGCATTCACAAACCAACTGTTAGGATTGAATCTATTATACCTGCTTTCTCAAAACCGAGTGGCCGAGTTTCACACAGAACTTGAGCGTCTTCCAGTTGATGTTATAAGAACAGATCTTTATATCAGACACCCGCTTGCTCTAGAACAGTATTTAATGGAGGGCAGCTACAACAAAATATTCTTAGCAAAGGGTAATGTGCCAGCAGAGAGCTACACCTTCTTCATGGATACACTTTTAGAAACAGTTCGTGGAGAGATTGCAGCTTGCATTGAGAAAGCTTACCACACAATCCCTTGTGCCGAAGCCGCTAGACGACTTAACTTGTCTTCTCAACAAGCAGTACTTGAGTATGGAAAGAAGCGTAACTGGAGGTTAGGTCCTGACAACTGCTACCGGTTCACAAGCGAAGAATTAACATCTGTGGCCTTCTCGGGCATTCTTCCATCTGCTGAACTGGCTCAACAGACCATTGAGTATGCCAGACATCTTGAGATGATTGTATAA
Protein
MATVKDVATLYQNLKTEWAKKPRKLDKCGELLNKIKIALTQLTFLPSNNVAANQKELILARDVLEIGAQWAVAVKDVKAFERYMSQLKCYYFDYKDHLPESAFTNQLLGLNLLYLLSQNRVAEFHTELERLPVDVIRTDLYIRHPLALEQYLMEGSYNKIFLAKGNVPAESYTFFMDTLLETVRGEIAACIEKAYHTIPCAEAARRLNLSSQQAVLEYGKKRNWRLGPDNCYRFTSEELTSVAFSGILPSAELAQQTIEYARHLEMIV
Summary
Description
Component of the 26S proteasome, a multiprotein complex involved in the ATP-dependent degradation of ubiquitinated proteins. This complex plays a key role in the maintenance of protein homeostasis by removing misfolded or damaged proteins, which could impair cellular functions, and by removing proteins whose functions are no longer required. Therefore, the proteasome participates in numerous cellular processes, including cell cycle progression, apoptosis, or DNA damage repair.
Subunit
Component of the 19S proteasome regulatory particle complex. The 26S proteasome consists of a 20S core particle (CP) and two 19S regulatory subunits (RP). The regulatory particle is made of a lid composed of 9 subunits including PSMD8, a base containing 6 ATPases and few additional components. Interacts with DDI2 (By similarity).
Similarity
Belongs to the proteasome subunit S14 family.
Keywords
Complete proteome
Isopeptide bond
Proteasome
Reference proteome
Ubl conjugation
Feature
chain 26S proteasome non-ATPase regulatory subunit 8
Uniprot
H9JGE8
A0A194RIJ1
I4DKA5
A0A212FHJ4
A0A1E1WJR7
A0A2H1WAV7
+ More
S4P3H8 A0A3S2N3L9 A0A2P8XL95 A0A2J7RLF3 A0A1B6MMZ6 A0A2R7X488 A0A067QUP6 A0A023F8V4 R4G8D6 M9W042 A0A069DRQ0 A0A224XHX4 A0A0A9XXZ0 A0A3B4D5Y9 A0A3S2PMX2 A2RV94 A0A1L8F911 A0A3B1IZZ0 Q6NU38 A0A3B3B5D9 E2C8I6 E2AVX2 A0A3Q3VUA1 A0A0V0G604 A0A1S3HD68 E9II76 A0A151IM23 A3KMH9 Q6NTJ8 A0A1L8F3Y1 A0A060YRV1 H2TMF9 A0A3B4WSB0 A0A3B4VF98 C1BGR0 C1BJM3 A0A195D815 A0A195ERH8 B5XAB2 A0A232FH51 A0A146WEV2 K7J594 V4BX33 Q05AY5 H2LAC7 A0A3P9HBJ0 Q4RRE0 A0A026WM63 A0A210QRL2 E3TFR8 A0A151X525 A0A3P9AK56 D2A2W4 A0A3Q3E6M3 Q3TW90 E3TBX3 A0A3Q2QZ26 A0A1W5AN99 A0A158NE08 A0A3L8DKU1 A0A195BRC9 A0A1W5B388 Q9CPS5 A0A0P6CFW9 F4X1S7 Q3B8P5 Q3TVY0 A0A3L7H7Q0 Q99JB5 H3B6U6 A0A0B7AZ06 C1BI19 K1QVR0 Q9CX56 Q3TI95 G5AMB7 A0A1U7R5B2 A0A151PK60 A0A0N0BEV9 F7ACH7 A0A3Q2C9F3 A0A3Q2TQ19 F1LMQ3 G3SUK7 A0A3B3SU68 V9L154 A0A2Y9E6K0 A0A2A3EM46 A0A088A0Z2 I3MMZ3 Q8BKP5 A0A091DH60 T1JGC0 A0A2K6V682
S4P3H8 A0A3S2N3L9 A0A2P8XL95 A0A2J7RLF3 A0A1B6MMZ6 A0A2R7X488 A0A067QUP6 A0A023F8V4 R4G8D6 M9W042 A0A069DRQ0 A0A224XHX4 A0A0A9XXZ0 A0A3B4D5Y9 A0A3S2PMX2 A2RV94 A0A1L8F911 A0A3B1IZZ0 Q6NU38 A0A3B3B5D9 E2C8I6 E2AVX2 A0A3Q3VUA1 A0A0V0G604 A0A1S3HD68 E9II76 A0A151IM23 A3KMH9 Q6NTJ8 A0A1L8F3Y1 A0A060YRV1 H2TMF9 A0A3B4WSB0 A0A3B4VF98 C1BGR0 C1BJM3 A0A195D815 A0A195ERH8 B5XAB2 A0A232FH51 A0A146WEV2 K7J594 V4BX33 Q05AY5 H2LAC7 A0A3P9HBJ0 Q4RRE0 A0A026WM63 A0A210QRL2 E3TFR8 A0A151X525 A0A3P9AK56 D2A2W4 A0A3Q3E6M3 Q3TW90 E3TBX3 A0A3Q2QZ26 A0A1W5AN99 A0A158NE08 A0A3L8DKU1 A0A195BRC9 A0A1W5B388 Q9CPS5 A0A0P6CFW9 F4X1S7 Q3B8P5 Q3TVY0 A0A3L7H7Q0 Q99JB5 H3B6U6 A0A0B7AZ06 C1BI19 K1QVR0 Q9CX56 Q3TI95 G5AMB7 A0A1U7R5B2 A0A151PK60 A0A0N0BEV9 F7ACH7 A0A3Q2C9F3 A0A3Q2TQ19 F1LMQ3 G3SUK7 A0A3B3SU68 V9L154 A0A2Y9E6K0 A0A2A3EM46 A0A088A0Z2 I3MMZ3 Q8BKP5 A0A091DH60 T1JGC0 A0A2K6V682
Pubmed
19121390
26354079
22651552
22118469
23622113
29403074
+ More
24845553 25474469 29136191 26334808 25401762 26823975 27762356 25329095 29451363 20798317 21282665 24755649 21551351 20433749 28648823 20075255 23254933 17554307 15496914 24508170 28812685 20634964 25069045 18362917 19820115 10349636 11042159 11076861 11217851 12466851 16141073 21347285 30249741 19468303 21183079 21719571 15489334 29704459 9215903 22992520 16141072 16857966 21993625 28071753 22293439 17495919 15057822 29398115 29240929 24402279
24845553 25474469 29136191 26334808 25401762 26823975 27762356 25329095 29451363 20798317 21282665 24755649 21551351 20433749 28648823 20075255 23254933 17554307 15496914 24508170 28812685 20634964 25069045 18362917 19820115 10349636 11042159 11076861 11217851 12466851 16141073 21347285 30249741 19468303 21183079 21719571 15489334 29704459 9215903 22992520 16141072 16857966 21993625 28071753 22293439 17495919 15057822 29398115 29240929 24402279
EMBL
BABH01033562
BABH01033563
KQ460154
KPJ17259.1
AK401723
KQ458880
+ More
BAM18345.1 KPJ04610.1 AGBW02008501 OWR53213.1 GDQN01003829 JAT87225.1 ODYU01007436 SOQ50211.1 GAIX01009497 JAA83063.1 RSAL01002136 RVE40572.1 PYGN01001796 PSN32765.1 NEVH01002690 PNF41663.1 GEBQ01002663 JAT37314.1 KK856672 PTY26230.1 KK852929 KDR13661.1 GBBI01000862 JAC17850.1 ACPB03001320 GAHY01001157 JAA76353.1 KC683804 QKKF02012754 AGJ84698.1 RZF43470.1 GBGD01002543 JAC86346.1 GFTR01004330 JAW12096.1 GBHO01018800 GBRD01016925 GDHC01005369 JAG24804.1 JAG48902.1 JAQ13260.1 CM012449 RVE65169.1 BC133237 AAI33238.1 CM004480 OCT68074.1 BC068763 BC097621 BC106280 AAH68763.1 AAH97621.1 AAI06281.1 GL453666 EFN75716.1 GL443213 EFN62394.1 GECL01003399 JAP02725.1 GL763392 EFZ19728.1 KQ977074 KYN05938.1 BC131885 AAI31886.1 BC068963 AAH68963.1 CM004481 OCT66303.1 FR912386 CDQ91860.1 BT073789 ACO08213.1 BT074802 ACO09226.1 KQ981153 KYN09013.1 KQ981993 KYN30828.1 BT047981 BT058673 BT060388 ACI67782.1 ACN10386.1 ACN12748.1 NNAY01000239 OXU29788.1 GCES01057780 JAR28543.1 AAZX01000332 KB201891 ESO93619.1 BC123112 AAI23113.1 CAAE01015002 CAG09042.1 KK107152 EZA57053.1 NEDP02002277 OWF51361.1 GU589202 ADO29154.1 KQ982548 KYQ55318.1 KQ971338 EFA02229.1 AK159795 BAE35376.1 GU587850 ADO27809.1 ADTU01013085 QOIP01000007 RLU20458.1 KQ976424 KYM88514.1 AC164564 AK002921 AK003436 AK027973 AK150493 AK150982 AK159216 AK166718 BAB22458.2 BAB22789.2 BAC25683.1 BAE29609.1 GDIP01002590 JAN01126.1 GL888551 EGI59584.1 BC105894 AAI05895.1 AK159926 BAE35488.1 RAZU01000274 RLQ61351.1 BC004075 BC005717 AAH04075.1 AAH05717.1 AFYH01009887 AFYH01009888 HACG01039118 CEK85983.1 BT074248 ACO08672.1 JH818228 EKC33025.1 AK020132 AK167948 BAE39951.1 JH165993 EHA98177.1 AKHW03000044 KYO49145.1 KQ435821 KOX72468.1 AC118147 JW872507 AFP05025.1 KZ288215 PBC32574.1 AGTP01108620 AK051257 BAC34576.1 KN122579 KFO29605.1 JH432201
BAM18345.1 KPJ04610.1 AGBW02008501 OWR53213.1 GDQN01003829 JAT87225.1 ODYU01007436 SOQ50211.1 GAIX01009497 JAA83063.1 RSAL01002136 RVE40572.1 PYGN01001796 PSN32765.1 NEVH01002690 PNF41663.1 GEBQ01002663 JAT37314.1 KK856672 PTY26230.1 KK852929 KDR13661.1 GBBI01000862 JAC17850.1 ACPB03001320 GAHY01001157 JAA76353.1 KC683804 QKKF02012754 AGJ84698.1 RZF43470.1 GBGD01002543 JAC86346.1 GFTR01004330 JAW12096.1 GBHO01018800 GBRD01016925 GDHC01005369 JAG24804.1 JAG48902.1 JAQ13260.1 CM012449 RVE65169.1 BC133237 AAI33238.1 CM004480 OCT68074.1 BC068763 BC097621 BC106280 AAH68763.1 AAH97621.1 AAI06281.1 GL453666 EFN75716.1 GL443213 EFN62394.1 GECL01003399 JAP02725.1 GL763392 EFZ19728.1 KQ977074 KYN05938.1 BC131885 AAI31886.1 BC068963 AAH68963.1 CM004481 OCT66303.1 FR912386 CDQ91860.1 BT073789 ACO08213.1 BT074802 ACO09226.1 KQ981153 KYN09013.1 KQ981993 KYN30828.1 BT047981 BT058673 BT060388 ACI67782.1 ACN10386.1 ACN12748.1 NNAY01000239 OXU29788.1 GCES01057780 JAR28543.1 AAZX01000332 KB201891 ESO93619.1 BC123112 AAI23113.1 CAAE01015002 CAG09042.1 KK107152 EZA57053.1 NEDP02002277 OWF51361.1 GU589202 ADO29154.1 KQ982548 KYQ55318.1 KQ971338 EFA02229.1 AK159795 BAE35376.1 GU587850 ADO27809.1 ADTU01013085 QOIP01000007 RLU20458.1 KQ976424 KYM88514.1 AC164564 AK002921 AK003436 AK027973 AK150493 AK150982 AK159216 AK166718 BAB22458.2 BAB22789.2 BAC25683.1 BAE29609.1 GDIP01002590 JAN01126.1 GL888551 EGI59584.1 BC105894 AAI05895.1 AK159926 BAE35488.1 RAZU01000274 RLQ61351.1 BC004075 BC005717 AAH04075.1 AAH05717.1 AFYH01009887 AFYH01009888 HACG01039118 CEK85983.1 BT074248 ACO08672.1 JH818228 EKC33025.1 AK020132 AK167948 BAE39951.1 JH165993 EHA98177.1 AKHW03000044 KYO49145.1 KQ435821 KOX72468.1 AC118147 JW872507 AFP05025.1 KZ288215 PBC32574.1 AGTP01108620 AK051257 BAC34576.1 KN122579 KFO29605.1 JH432201
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000283053
UP000245037
+ More
UP000235965 UP000027135 UP000015103 UP000291343 UP000261440 UP000186698 UP000018467 UP000261560 UP000008237 UP000000311 UP000261620 UP000085678 UP000078542 UP000193380 UP000005226 UP000261360 UP000261420 UP000078492 UP000078541 UP000087266 UP000215335 UP000002358 UP000030746 UP000001038 UP000265180 UP000265200 UP000007303 UP000053097 UP000242188 UP000221080 UP000075809 UP000265140 UP000007266 UP000264820 UP000265000 UP000192224 UP000005205 UP000279307 UP000078540 UP000000589 UP000007755 UP000273346 UP000008672 UP000005408 UP000006813 UP000189706 UP000050525 UP000053105 UP000002280 UP000265020 UP000002494 UP000007646 UP000261540 UP000248480 UP000242457 UP000005203 UP000005215 UP000028990 UP000233220
UP000235965 UP000027135 UP000015103 UP000291343 UP000261440 UP000186698 UP000018467 UP000261560 UP000008237 UP000000311 UP000261620 UP000085678 UP000078542 UP000193380 UP000005226 UP000261360 UP000261420 UP000078492 UP000078541 UP000087266 UP000215335 UP000002358 UP000030746 UP000001038 UP000265180 UP000265200 UP000007303 UP000053097 UP000242188 UP000221080 UP000075809 UP000265140 UP000007266 UP000264820 UP000265000 UP000192224 UP000005205 UP000279307 UP000078540 UP000000589 UP000007755 UP000273346 UP000008672 UP000005408 UP000006813 UP000189706 UP000050525 UP000053105 UP000002280 UP000265020 UP000002494 UP000007646 UP000261540 UP000248480 UP000242457 UP000005203 UP000005215 UP000028990 UP000233220
Pfam
PF10075 CSN8_PSD8_EIF3K
Interpro
ProteinModelPortal
H9JGE8
A0A194RIJ1
I4DKA5
A0A212FHJ4
A0A1E1WJR7
A0A2H1WAV7
+ More
S4P3H8 A0A3S2N3L9 A0A2P8XL95 A0A2J7RLF3 A0A1B6MMZ6 A0A2R7X488 A0A067QUP6 A0A023F8V4 R4G8D6 M9W042 A0A069DRQ0 A0A224XHX4 A0A0A9XXZ0 A0A3B4D5Y9 A0A3S2PMX2 A2RV94 A0A1L8F911 A0A3B1IZZ0 Q6NU38 A0A3B3B5D9 E2C8I6 E2AVX2 A0A3Q3VUA1 A0A0V0G604 A0A1S3HD68 E9II76 A0A151IM23 A3KMH9 Q6NTJ8 A0A1L8F3Y1 A0A060YRV1 H2TMF9 A0A3B4WSB0 A0A3B4VF98 C1BGR0 C1BJM3 A0A195D815 A0A195ERH8 B5XAB2 A0A232FH51 A0A146WEV2 K7J594 V4BX33 Q05AY5 H2LAC7 A0A3P9HBJ0 Q4RRE0 A0A026WM63 A0A210QRL2 E3TFR8 A0A151X525 A0A3P9AK56 D2A2W4 A0A3Q3E6M3 Q3TW90 E3TBX3 A0A3Q2QZ26 A0A1W5AN99 A0A158NE08 A0A3L8DKU1 A0A195BRC9 A0A1W5B388 Q9CPS5 A0A0P6CFW9 F4X1S7 Q3B8P5 Q3TVY0 A0A3L7H7Q0 Q99JB5 H3B6U6 A0A0B7AZ06 C1BI19 K1QVR0 Q9CX56 Q3TI95 G5AMB7 A0A1U7R5B2 A0A151PK60 A0A0N0BEV9 F7ACH7 A0A3Q2C9F3 A0A3Q2TQ19 F1LMQ3 G3SUK7 A0A3B3SU68 V9L154 A0A2Y9E6K0 A0A2A3EM46 A0A088A0Z2 I3MMZ3 Q8BKP5 A0A091DH60 T1JGC0 A0A2K6V682
S4P3H8 A0A3S2N3L9 A0A2P8XL95 A0A2J7RLF3 A0A1B6MMZ6 A0A2R7X488 A0A067QUP6 A0A023F8V4 R4G8D6 M9W042 A0A069DRQ0 A0A224XHX4 A0A0A9XXZ0 A0A3B4D5Y9 A0A3S2PMX2 A2RV94 A0A1L8F911 A0A3B1IZZ0 Q6NU38 A0A3B3B5D9 E2C8I6 E2AVX2 A0A3Q3VUA1 A0A0V0G604 A0A1S3HD68 E9II76 A0A151IM23 A3KMH9 Q6NTJ8 A0A1L8F3Y1 A0A060YRV1 H2TMF9 A0A3B4WSB0 A0A3B4VF98 C1BGR0 C1BJM3 A0A195D815 A0A195ERH8 B5XAB2 A0A232FH51 A0A146WEV2 K7J594 V4BX33 Q05AY5 H2LAC7 A0A3P9HBJ0 Q4RRE0 A0A026WM63 A0A210QRL2 E3TFR8 A0A151X525 A0A3P9AK56 D2A2W4 A0A3Q3E6M3 Q3TW90 E3TBX3 A0A3Q2QZ26 A0A1W5AN99 A0A158NE08 A0A3L8DKU1 A0A195BRC9 A0A1W5B388 Q9CPS5 A0A0P6CFW9 F4X1S7 Q3B8P5 Q3TVY0 A0A3L7H7Q0 Q99JB5 H3B6U6 A0A0B7AZ06 C1BI19 K1QVR0 Q9CX56 Q3TI95 G5AMB7 A0A1U7R5B2 A0A151PK60 A0A0N0BEV9 F7ACH7 A0A3Q2C9F3 A0A3Q2TQ19 F1LMQ3 G3SUK7 A0A3B3SU68 V9L154 A0A2Y9E6K0 A0A2A3EM46 A0A088A0Z2 I3MMZ3 Q8BKP5 A0A091DH60 T1JGC0 A0A2K6V682
PDB
6EPF
E-value=2.67376e-86,
Score=810
Ontologies
PANTHER
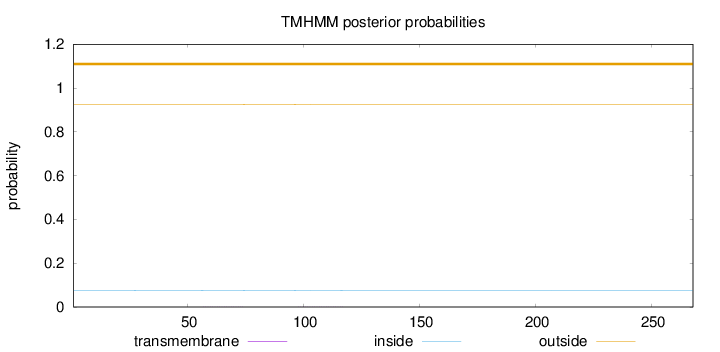
Topology
Length:
268
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01543
Exp number, first 60 AAs:
0.00306
Total prob of N-in:
0.07479
outside
1 - 268
Population Genetic Test Statistics
Pi
13.97499
Theta
17.344907
Tajima's D
-0.479801
CLR
1.835403
CSRT
0.246087695615219
Interpretation
Uncertain
