Gene
KWMTBOMO11160 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008596
Annotation
PREDICTED:_nuclear_valosin-containing_protein-like_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.504
Sequence
CDS
ATGGCGCGTTCTCTGAAAGAGCGGTACCGCGAGTATAACAATAGGAGCGACACTGCCTTCTGTCAGATGGTCGAAGCTGCGTACAAGGTTGTACTACAGAGTTACGGCCTCGACGGAGCGTCCACATCAGATGGGTCGGAGGAAGAGTCAGATTTAGAATTGTTAGACGAAAACACCAGTGCGTTAAACGAACGTATATTGGCCATGTACAAAATGCAAGATAAAAAACGTCCGGCTGCGAGTCGGCGCCCCACCGACAAGGCAGGCGAGCCTATCGACATCTTGAGCAGCGATGAAGACGGCAGTGCGGCTCCCAAATTACCTAAAATTAACGACCAAATAACTATCACACCACATATCCCAGCCAGAACTAACAGCACGGCGATTGAGAGTGCACTCAAACAACAAAACCAACCACTTGATAAAAAACGTAAGCTTGACAGCAATAAAAGTGGAGCACCGAAGAAAAAACATGAGACTGGATCAGCAAAAATGGTTAAAATAGGTTTTGAGGATATTGGTGGTCTTTCTGACATCATTACCCAGATCTGCGACTTGATATTACACATGAAACACCCTGAAGTATATAGTAAATTGGGAATTAAAGCACCCAGAGGAGCATTACTACATGGCCCTCCGGGGACGGGTAAAACCTTATTAGCACATGCAATAGCAGGTAAACTGCAGTTACCTCTTGTTTCTGTGACTGGAACTGAATTAGTTGGAGGTGTGTCTGGCGAGTCGGAAGAGAGAATTAGAGAGTTGTTTGAGAGAGCTCAAGCTGCCGCACCAAGTGTGTTGTTTATTGATGAAATTGATGCAGTTTGTGGTAACCGAATACATGCACAAAAAGACATGGAAAAACGTATGGTTGCTCAATTACTGGCTAGTTTAGATAGTTTGAGTGAAAATAACTCTTCAGTTTTAGTTCTTGCAGCTACAAATAACCCTGATGCCTTAGATCCTGCTTTGAGAAGGGCAGGTCGCCTAGAGCAAGAGATTACTTTAGGTATCCCTACATTAAAGGCTAGGAAAGAAATAATAAACATTCTCTGTAAAAACATAACGTTGAGTGAAGATGTGGACATGAACATTTTAGCTCAAGTCACTCCCGGTTTCGTTGGAGCTGATTTACAAGCTCTGGTGAACAAAGCTAGCACATATGCAGTGAAAAGGATATTTAGTGAAATTCATGCAGAAGAATTAAAAGCTAGTAAAGAAGTCATTAAAGTTGATTCAAATAATGATGAGATCGCTATTGTATCAAATGGTGAAAAAGAAAGTGAAGACAAAAATAAAGAAAATGAGACAACGGATGATGTAGCTAAACCTACAGAGGAGCAGCTACCTATTGAACCAATAACTAATGAGGCAAACAAGGATATTACAGAAGGACAGGAAGAGAATGTAAGCAGTAACCCTCCAGAAAAACTTAGCATTGTTGATGAAGTTTTAGAGCTTCTCAATGTCAGGCCATACAGCGTAGACAAATTATCAGGCCTTGCCATTAAACAAGAAGATTTTTTGAAAGCTTTAAAAACCACAAAACCTTGTGCTGTAAGGGAGGGTATTGTGACTGTCCCAGATGTAACATGGGATGATGTGGGGTCGTTAAAGCAAGTCCGCAAGGATTTACAACTGGCTGTACTTGGTCCAGTTAAATACCCAGAACTCTTAAACAGCTTAGGACTGTCTGCGGCGAGCGGTGTGCTTCTTTGTGGTCCACCAGGATGCGGTAAAACTTTGCTGGCAAAAGCAGTGGCCAATGAAGCTGGCGTTAATTTTATCTCAGTTAAGGGTCCTGAGCTGCTCAATATGTACGTGGGTGAGAGCGAGCGCGCCGTCAGGACTTGCTTCCGTCGAGCTCGGAACTCGGCACCCTGCGTTATATTCTTTGATGAGTTTGATGCGCTCTGTCCGCGACGAACGTCACGCGATCACGACGGTGCGGCTCGCGTCGTCAACCAGCTCCTTACTGAGATGGACGGCATCGAAGGGCGCGAGGGCGTGTTCGTACTCGCAGCCTCCAACCGACCGGACATTATAGATCCCGCCGTACTGCGGCCCGGCAGACTCGACCGCGTTATGTACGTCGGCATGCCGGCCCGGGACGATCGAAGCGATATCCTCGTCAAGCTCACCAAGGGAGGATCTCGGCCGCGCCTGGCTGCCGACGTGGACCTGCACGAGGTGGCGGGGCTCACCGACGGGTACACAGGTGCCGACCTGGGCGGACTGGTGCGGACTGCGGCCACGCTAGCATTGAGCGAGCTCATCGCCGAGGGTCGCGGGGCGGACGCCGACGTGTCGGTGCGCGCCGAGCACTTTCGTGCGGCCCTAGCGCGCTGCAAACCTTCCGTTTCGGCGAAGGAGCAGCTTCATTACGAGAAGCTAAGGATGAAATACGCATCTGGAGCAGACGACAGGGACATGGAGATGGAGACGGAGCCGCTGGTGGAGGAGTCGATGGACACCGTGTGA
Protein
MARSLKERYREYNNRSDTAFCQMVEAAYKVVLQSYGLDGASTSDGSEEESDLELLDENTSALNERILAMYKMQDKKRPAASRRPTDKAGEPIDILSSDEDGSAAPKLPKINDQITITPHIPARTNSTAIESALKQQNQPLDKKRKLDSNKSGAPKKKHETGSAKMVKIGFEDIGGLSDIITQICDLILHMKHPEVYSKLGIKAPRGALLHGPPGTGKTLLAHAIAGKLQLPLVSVTGTELVGGVSGESEERIRELFERAQAAAPSVLFIDEIDAVCGNRIHAQKDMEKRMVAQLLASLDSLSENNSSVLVLAATNNPDALDPALRRAGRLEQEITLGIPTLKARKEIINILCKNITLSEDVDMNILAQVTPGFVGADLQALVNKASTYAVKRIFSEIHAEELKASKEVIKVDSNNDEIAIVSNGEKESEDKNKENETTDDVAKPTEEQLPIEPITNEANKDITEGQEENVSSNPPEKLSIVDEVLELLNVRPYSVDKLSGLAIKQEDFLKALKTTKPCAVREGIVTVPDVTWDDVGSLKQVRKDLQLAVLGPVKYPELLNSLGLSAASGVLLCGPPGCGKTLLAKAVANEAGVNFISVKGPELLNMYVGESERAVRTCFRRARNSAPCVIFFDEFDALCPRRTSRDHDGAARVVNQLLTEMDGIEGREGVFVLAASNRPDIIDPAVLRPGRLDRVMYVGMPARDDRSDILVKLTKGGSRPRLAADVDLHEVAGLTDGYTGADLGGLVRTAATLALSELIAEGRGADADVSVRAEHFRAALARCKPSVSAKEQLHYEKLRMKYASGADDRDMEMETEPLVEESMDTV
Summary
Similarity
Belongs to the AAA ATPase family.
Uniprot
H9JGE9
A0A2H1WAY0
A0A0L7LPN4
A0A194RIU7
A0A194QMA8
S4PKJ2
+ More
A0A1B0A6B9 A0A1J1J7B2 D6WIP4 A0A1Y1NHJ3 A0A1A9VKY3 A0A1B0FKC4 A0A1A9XE84 A0A1A9WD13 A0A1B0C645 T1PHP6 A0A1B0FIN4 E2BIR9 A0A0L0CPZ6 A0A2P8XIU3 A0A3B0J5R6 A0A1B6H1Q9 A0A158NNN5 A0A1B6E946 A0A1B6EFC1 A0A1B6FLR1 A0A067RX51 A0A2J7QDC7 A0A026WRY0 A0A3B0K1B4 F4WEY0 E2AYL6 A0A210QD30 B4LDW8 B4KYP6 A0A3B4UTL0 A0A3Q3MG76 W5UJS9 A0A3Q3JJD6 A0A3B4CRI0 W5KPH2 I3K5Y1 A0A2I0M4Z9 A0A099ZE11 I3K5Y2 A0A1W7RF74 A0A224XJW8 A0A3Q3FII4 A0A1D2NAJ0 A0A2R7WUL1 A0A3Q1IXV5 A0A0F8B0D7 A0A1B6F5C6 A0A1X2GRL6 A0A093F1M7 A0A3B4E8J4 A0A0M4EG45 A0A1C7N6G7 A0A0C7BK78 A0A0C7BKS2 A0A168PG17 A0A1X0S0R7 A0A097F6Y2 A0A068S844 A0A367JCG8 F7E892 A0A0C7CAE8 A0A1X0QU86 A0A0C9MAV6 A0A2G4SJE4 T1HCW5 T1H4K0 S2IZN9 A0A077WB31 A0A1X2I572 A0A163J6D5 I1BS28 A0A0M9VR06 A0A0B7N1E5 A0A0L0H741 A0A165E2W4 A0A1Y1WCP8 A0A316YY03 A0A1Y2GYU8 A0A1Y1YIW0 A0A316VDL1 A0A1B9GG11 A0A397TS93 B6K792 A0A3N4M691 A0A1B9IH40 A0A225WI79 A0A369JV61 A0A1E4RRW9 A0A1M7ZZR6 M5E7L7 B5VMR2 A0A1Y2FEE9
A0A1B0A6B9 A0A1J1J7B2 D6WIP4 A0A1Y1NHJ3 A0A1A9VKY3 A0A1B0FKC4 A0A1A9XE84 A0A1A9WD13 A0A1B0C645 T1PHP6 A0A1B0FIN4 E2BIR9 A0A0L0CPZ6 A0A2P8XIU3 A0A3B0J5R6 A0A1B6H1Q9 A0A158NNN5 A0A1B6E946 A0A1B6EFC1 A0A1B6FLR1 A0A067RX51 A0A2J7QDC7 A0A026WRY0 A0A3B0K1B4 F4WEY0 E2AYL6 A0A210QD30 B4LDW8 B4KYP6 A0A3B4UTL0 A0A3Q3MG76 W5UJS9 A0A3Q3JJD6 A0A3B4CRI0 W5KPH2 I3K5Y1 A0A2I0M4Z9 A0A099ZE11 I3K5Y2 A0A1W7RF74 A0A224XJW8 A0A3Q3FII4 A0A1D2NAJ0 A0A2R7WUL1 A0A3Q1IXV5 A0A0F8B0D7 A0A1B6F5C6 A0A1X2GRL6 A0A093F1M7 A0A3B4E8J4 A0A0M4EG45 A0A1C7N6G7 A0A0C7BK78 A0A0C7BKS2 A0A168PG17 A0A1X0S0R7 A0A097F6Y2 A0A068S844 A0A367JCG8 F7E892 A0A0C7CAE8 A0A1X0QU86 A0A0C9MAV6 A0A2G4SJE4 T1HCW5 T1H4K0 S2IZN9 A0A077WB31 A0A1X2I572 A0A163J6D5 I1BS28 A0A0M9VR06 A0A0B7N1E5 A0A0L0H741 A0A165E2W4 A0A1Y1WCP8 A0A316YY03 A0A1Y2GYU8 A0A1Y1YIW0 A0A316VDL1 A0A1B9GG11 A0A397TS93 B6K792 A0A3N4M691 A0A1B9IH40 A0A225WI79 A0A369JV61 A0A1E4RRW9 A0A1M7ZZR6 M5E7L7 B5VMR2 A0A1Y2FEE9
Pubmed
19121390
26227816
26354079
23622113
18362917
19820115
+ More
28004739 25315136 20798317 26108605 29403074 21347285 24845553 24508170 30249741 21719571 28812685 17994087 23127152 25329095 25186727 23371554 26358130 27289101 25835551 27956601 25006230 29674435 17495919 19578406 26659563 29771364 21511999 30420746 18778279
28004739 25315136 20798317 26108605 29403074 21347285 24845553 24508170 30249741 21719571 28812685 17994087 23127152 25329095 25186727 23371554 26358130 27289101 25835551 27956601 25006230 29674435 17495919 19578406 26659563 29771364 21511999 30420746 18778279
EMBL
BABH01033563
BABH01033564
ODYU01007436
SOQ50213.1
JTDY01000391
KOB77397.1
+ More
KQ460154 KPJ17260.1 KQ458880 KPJ04611.1 GAIX01004465 JAA88095.1 CVRI01000075 CRL08317.1 KQ971321 EEZ99985.1 GEZM01002092 JAV97372.1 CCAG010004976 JXJN01026428 KA648204 AFP62833.1 CCAG010004424 GL448530 EFN84409.1 JRES01000080 KNC34336.1 PYGN01001996 PSN31892.1 OUUW01000002 SPP77177.1 GECZ01001191 JAS68578.1 ADTU01002754 GEDC01014982 GEDC01002839 JAS22316.1 JAS34459.1 GEDC01000683 JAS36615.1 GECZ01018659 JAS51110.1 KK852411 KDR24474.1 NEVH01015825 PNF26579.1 KK107119 QOIP01000009 EZA58782.1 RLU18328.1 SPP77178.1 GL888112 EGI67251.1 GL443930 EFN61467.1 NEDP02004116 OWF46639.1 CH940647 EDW68991.2 CH933809 EDW17760.2 JT417153 AHH42276.1 AERX01042071 AKCR02000039 PKK24759.1 KL891754 KGL79090.1 GDAY02001656 JAV49771.1 GFTR01008117 JAW08309.1 LJIJ01000118 ODN02269.1 KK855491 PTY22810.1 KQ042127 KKF18771.1 GECZ01024404 JAS45365.1 MCGT01000005 ORX59716.1 KK384550 KFV50375.1 CP012525 ALC43327.1 LUGH01000475 OBZ84743.1 CCYT01000020 CEG71680.1 CEG71681.1 AMYB01000001 OAD07683.1 KV921344 ORE17866.1 KJ999714 AIT18235.1 CBTN010000046 CDH57446.1 PJQM01003677 RCH87634.1 CCYT01000217 CEG80560.1 KV922009 ORE03309.1 DF836354 GAN04479.1 KZ303861 PHZ08881.1 ACPB03000292 ACPB03000293 CAQQ02180757 CAQQ02180758 KE124079 EPB83236.1 LK023313 CDS03348.1 MCGE01000027 ORZ09612.1 LT551811 SAL97443.1 CH476733 EIE79008.1 LGAV01000001 KOS16112.1 LN725930 CEP11252.1 KQ257467 KNC96686.1 KV427626 KZT06140.1 MCFD01000004 ORX71310.1 KZ819634 PWN94327.1 MCFF01000004 ORZ27446.1 MCFE01000123 ORX97915.1 KZ819603 PWN35414.1 KI894018 OCF29956.1 QKYT01000003 RIA99505.1 KE651168 EEB09396.2 ML121529 RPB28522.1 KI669469 OCF54670.1 NBNE01000754 OWZ17423.1 LUEZ02000040 RDB26229.1 KV454538 ODV70039.1 LT671821 SHO75685.1 HE999552 CCU98469.1 ABSV01001543 EDZ70783.1 MCOG01000010 ORY81686.1
KQ460154 KPJ17260.1 KQ458880 KPJ04611.1 GAIX01004465 JAA88095.1 CVRI01000075 CRL08317.1 KQ971321 EEZ99985.1 GEZM01002092 JAV97372.1 CCAG010004976 JXJN01026428 KA648204 AFP62833.1 CCAG010004424 GL448530 EFN84409.1 JRES01000080 KNC34336.1 PYGN01001996 PSN31892.1 OUUW01000002 SPP77177.1 GECZ01001191 JAS68578.1 ADTU01002754 GEDC01014982 GEDC01002839 JAS22316.1 JAS34459.1 GEDC01000683 JAS36615.1 GECZ01018659 JAS51110.1 KK852411 KDR24474.1 NEVH01015825 PNF26579.1 KK107119 QOIP01000009 EZA58782.1 RLU18328.1 SPP77178.1 GL888112 EGI67251.1 GL443930 EFN61467.1 NEDP02004116 OWF46639.1 CH940647 EDW68991.2 CH933809 EDW17760.2 JT417153 AHH42276.1 AERX01042071 AKCR02000039 PKK24759.1 KL891754 KGL79090.1 GDAY02001656 JAV49771.1 GFTR01008117 JAW08309.1 LJIJ01000118 ODN02269.1 KK855491 PTY22810.1 KQ042127 KKF18771.1 GECZ01024404 JAS45365.1 MCGT01000005 ORX59716.1 KK384550 KFV50375.1 CP012525 ALC43327.1 LUGH01000475 OBZ84743.1 CCYT01000020 CEG71680.1 CEG71681.1 AMYB01000001 OAD07683.1 KV921344 ORE17866.1 KJ999714 AIT18235.1 CBTN010000046 CDH57446.1 PJQM01003677 RCH87634.1 CCYT01000217 CEG80560.1 KV922009 ORE03309.1 DF836354 GAN04479.1 KZ303861 PHZ08881.1 ACPB03000292 ACPB03000293 CAQQ02180757 CAQQ02180758 KE124079 EPB83236.1 LK023313 CDS03348.1 MCGE01000027 ORZ09612.1 LT551811 SAL97443.1 CH476733 EIE79008.1 LGAV01000001 KOS16112.1 LN725930 CEP11252.1 KQ257467 KNC96686.1 KV427626 KZT06140.1 MCFD01000004 ORX71310.1 KZ819634 PWN94327.1 MCFF01000004 ORZ27446.1 MCFE01000123 ORX97915.1 KZ819603 PWN35414.1 KI894018 OCF29956.1 QKYT01000003 RIA99505.1 KE651168 EEB09396.2 ML121529 RPB28522.1 KI669469 OCF54670.1 NBNE01000754 OWZ17423.1 LUEZ02000040 RDB26229.1 KV454538 ODV70039.1 LT671821 SHO75685.1 HE999552 CCU98469.1 ABSV01001543 EDZ70783.1 MCOG01000010 ORY81686.1
Proteomes
UP000005204
UP000037510
UP000053240
UP000053268
UP000092445
UP000183832
+ More
UP000007266 UP000078200 UP000092444 UP000092443 UP000091820 UP000092460 UP000095301 UP000008237 UP000037069 UP000245037 UP000268350 UP000005205 UP000027135 UP000235965 UP000053097 UP000279307 UP000007755 UP000000311 UP000242188 UP000008792 UP000009192 UP000261420 UP000261660 UP000221080 UP000261600 UP000261440 UP000018467 UP000005207 UP000053872 UP000053641 UP000094527 UP000265040 UP000242146 UP000092553 UP000093000 UP000054919 UP000077051 UP000242381 UP000027586 UP000253551 UP000002280 UP000242414 UP000053815 UP000242254 UP000015103 UP000015102 UP000014254 UP000193560 UP000078561 UP000009138 UP000037751 UP000054107 UP000053201 UP000076871 UP000193922 UP000245768 UP000193648 UP000193498 UP000245771 UP000092730 UP000265703 UP000001744 UP000267821 UP000092583 UP000198211 UP000076154 UP000095085 UP000186303 UP000008988 UP000193920
UP000007266 UP000078200 UP000092444 UP000092443 UP000091820 UP000092460 UP000095301 UP000008237 UP000037069 UP000245037 UP000268350 UP000005205 UP000027135 UP000235965 UP000053097 UP000279307 UP000007755 UP000000311 UP000242188 UP000008792 UP000009192 UP000261420 UP000261660 UP000221080 UP000261600 UP000261440 UP000018467 UP000005207 UP000053872 UP000053641 UP000094527 UP000265040 UP000242146 UP000092553 UP000093000 UP000054919 UP000077051 UP000242381 UP000027586 UP000253551 UP000002280 UP000242414 UP000053815 UP000242254 UP000015103 UP000015102 UP000014254 UP000193560 UP000078561 UP000009138 UP000037751 UP000054107 UP000053201 UP000076871 UP000193922 UP000245768 UP000193648 UP000193498 UP000245771 UP000092730 UP000265703 UP000001744 UP000267821 UP000092583 UP000198211 UP000076154 UP000095085 UP000186303 UP000008988 UP000193920
PRIDE
Pfam
Interpro
SUPFAM
SSF52540
SSF52540
Gene 3D
ProteinModelPortal
H9JGE9
A0A2H1WAY0
A0A0L7LPN4
A0A194RIU7
A0A194QMA8
S4PKJ2
+ More
A0A1B0A6B9 A0A1J1J7B2 D6WIP4 A0A1Y1NHJ3 A0A1A9VKY3 A0A1B0FKC4 A0A1A9XE84 A0A1A9WD13 A0A1B0C645 T1PHP6 A0A1B0FIN4 E2BIR9 A0A0L0CPZ6 A0A2P8XIU3 A0A3B0J5R6 A0A1B6H1Q9 A0A158NNN5 A0A1B6E946 A0A1B6EFC1 A0A1B6FLR1 A0A067RX51 A0A2J7QDC7 A0A026WRY0 A0A3B0K1B4 F4WEY0 E2AYL6 A0A210QD30 B4LDW8 B4KYP6 A0A3B4UTL0 A0A3Q3MG76 W5UJS9 A0A3Q3JJD6 A0A3B4CRI0 W5KPH2 I3K5Y1 A0A2I0M4Z9 A0A099ZE11 I3K5Y2 A0A1W7RF74 A0A224XJW8 A0A3Q3FII4 A0A1D2NAJ0 A0A2R7WUL1 A0A3Q1IXV5 A0A0F8B0D7 A0A1B6F5C6 A0A1X2GRL6 A0A093F1M7 A0A3B4E8J4 A0A0M4EG45 A0A1C7N6G7 A0A0C7BK78 A0A0C7BKS2 A0A168PG17 A0A1X0S0R7 A0A097F6Y2 A0A068S844 A0A367JCG8 F7E892 A0A0C7CAE8 A0A1X0QU86 A0A0C9MAV6 A0A2G4SJE4 T1HCW5 T1H4K0 S2IZN9 A0A077WB31 A0A1X2I572 A0A163J6D5 I1BS28 A0A0M9VR06 A0A0B7N1E5 A0A0L0H741 A0A165E2W4 A0A1Y1WCP8 A0A316YY03 A0A1Y2GYU8 A0A1Y1YIW0 A0A316VDL1 A0A1B9GG11 A0A397TS93 B6K792 A0A3N4M691 A0A1B9IH40 A0A225WI79 A0A369JV61 A0A1E4RRW9 A0A1M7ZZR6 M5E7L7 B5VMR2 A0A1Y2FEE9
A0A1B0A6B9 A0A1J1J7B2 D6WIP4 A0A1Y1NHJ3 A0A1A9VKY3 A0A1B0FKC4 A0A1A9XE84 A0A1A9WD13 A0A1B0C645 T1PHP6 A0A1B0FIN4 E2BIR9 A0A0L0CPZ6 A0A2P8XIU3 A0A3B0J5R6 A0A1B6H1Q9 A0A158NNN5 A0A1B6E946 A0A1B6EFC1 A0A1B6FLR1 A0A067RX51 A0A2J7QDC7 A0A026WRY0 A0A3B0K1B4 F4WEY0 E2AYL6 A0A210QD30 B4LDW8 B4KYP6 A0A3B4UTL0 A0A3Q3MG76 W5UJS9 A0A3Q3JJD6 A0A3B4CRI0 W5KPH2 I3K5Y1 A0A2I0M4Z9 A0A099ZE11 I3K5Y2 A0A1W7RF74 A0A224XJW8 A0A3Q3FII4 A0A1D2NAJ0 A0A2R7WUL1 A0A3Q1IXV5 A0A0F8B0D7 A0A1B6F5C6 A0A1X2GRL6 A0A093F1M7 A0A3B4E8J4 A0A0M4EG45 A0A1C7N6G7 A0A0C7BK78 A0A0C7BKS2 A0A168PG17 A0A1X0S0R7 A0A097F6Y2 A0A068S844 A0A367JCG8 F7E892 A0A0C7CAE8 A0A1X0QU86 A0A0C9MAV6 A0A2G4SJE4 T1HCW5 T1H4K0 S2IZN9 A0A077WB31 A0A1X2I572 A0A163J6D5 I1BS28 A0A0M9VR06 A0A0B7N1E5 A0A0L0H741 A0A165E2W4 A0A1Y1WCP8 A0A316YY03 A0A1Y2GYU8 A0A1Y1YIW0 A0A316VDL1 A0A1B9GG11 A0A397TS93 B6K792 A0A3N4M691 A0A1B9IH40 A0A225WI79 A0A369JV61 A0A1E4RRW9 A0A1M7ZZR6 M5E7L7 B5VMR2 A0A1Y2FEE9
PDB
6MAT
E-value=1.01722e-95,
Score=897
Ontologies
GO
GO:0005524
GO:0005201
GO:0016887
GO:0051973
GO:0042254
GO:0005634
GO:1990275
GO:0004181
GO:0008270
GO:0042273
GO:0005730
GO:0005654
GO:0032092
GO:0006364
GO:1904749
GO:0000176
GO:0005697
GO:0016787
GO:0000055
GO:0030687
GO:0006468
GO:0007169
GO:0016020
GO:0005515
GO:0006811
GO:0005622
GO:0006810
GO:0003824
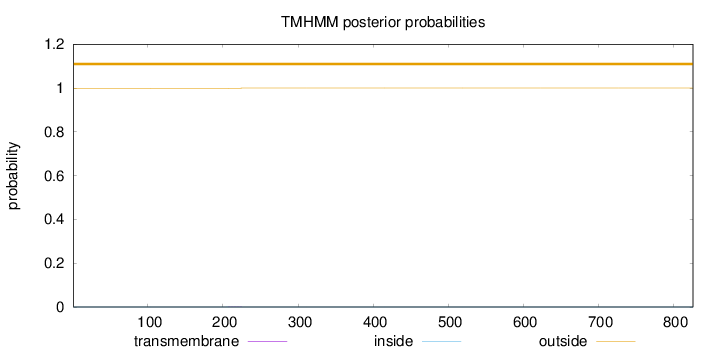
Topology
Length:
826
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00802999999999997
Exp number, first 60 AAs:
0.00142
Total prob of N-in:
0.00054
outside
1 - 826
Population Genetic Test Statistics
Pi
5.237239
Theta
7.499751
Tajima's D
-1.212859
CLR
12.685546
CSRT
0.0971451427428629
Interpretation
Uncertain
