Gene
KWMTBOMO11159
Pre Gene Modal
BGIBMGA008587
Annotation
PREDICTED:_uncharacterized_protein_K02A2.6-like_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 2.718
Sequence
CDS
ATGTCACTTATATCAAATTTGAAATCCTTTGATGGAGAAAAGTGGTCAGTGTTCCAGGAACAGTTAGACTGTTTCTTTATCGTTAATGAAGTGTCCGATGAAAAAAAAGTACCACTATTAATCACTAAATTAGCACCCAAAGTTTATGAAGTCCTCTCTTATTTATGTAAACCAAAAAAACCACAGGATCTAAAATTTGACCAACTTTGTTTCAAGTTAGCAGCAAAATATGATCAAAAACTACCATCAGCCATTGAAAGAGCAAATTTCAGACAGCGGAACCAGGCACCGAATGAAAAAGTAGAAGATTACGTCTTGGAACTGAGGAAACTTGCTGCAAAATGTAACTTCAAAGATATGGAGGACCAAGTCAAAGAGAAACTAATGGACGGAGTGTCCTCAAAAATGATAAAGTTTGAATTGATGAAGAGTGCTGGTGATGCAACACTAGAAAGGTGTATTGAATTAGCCCGCACTGTTGAAGCAGCAATATTACAAACAAACGGGGCCACAAATGAAGTTGCGGAAATATTTGTCAGCAAATCAACCAATAAATCACCACAACAGCAAAGAAATACCTACCAACAAAAAAATCAATGTTACTGCTGCGGAAAATGGAACCACCTAAAGCGGGACTGCAGACTCAATAGAAAATATTGCTCTGAATGTGGAATTCAAGGACACATCTTCAGAATGTGTCCCAAGAAGCAAAGAAAAGTAAATGTTCTGCAGGAAGATAACCAACAAAAGGATAAAAATACAGATAATGAACCCACTGAGTTTGAACCAAAATATTATGAAACATATGCATGTAACACTGTGAGTAAAATACCACCTTACTACATAACCATAAAAATGGAGGAGTGTAAGCTACCATTTCAACTAGACACCGGCTCAGAAGTGACAGTAATCACGGAAAATGACAAAAATGACATTTTAATAAACCACGTACTGCAAGATTGTGACATAATATTTAAAAATTTCGATCAATCCACAACAAGGCCACTAGGCATAGTTCATAATATATCTGTAGAATATGGTAACAGAAGTGTTTACTTAAAAGCTTTTGTAGTAAAAAATAAGTCACCCCGCATAATAGGCAGAGATTGGCTAAACGCCTTAGATGCACAAAATAGTATAGAAAAAAAATTCGCAGCAGTATTCAGCCCAGGCTGGGGAAATTTCAAAGGCGATGTCATAAATTTAGTTTTGAAAAAGGAGGTAGAACCCAAGTGTTTACCAGTGAGACGCGTACCATATGCTCTTAAGGACAAAGTAAATAATGAAATAAAAAGATTATTGGACAATGGTAGGATCACAGTAGTAAAACACAGTAAATGGGGCACCCCTGTAGTACCAATTTTAAAATCGGATGGTTCAGTAAGATTGTGTGGAGATTACAAGTTAACAGTAAACAAGTGTTTGGAAGTAGATCATTTCCCTTTACCGCATGTTGAAGACATTATAAACACCCTACAAGGGGGCGAATATTACTGCGAATTGGACTTAAAAGAAGCATATTTGCAAGCGCCTTTAAGTTTAGAAAGCCAGGATTGCACAACCATTGTAACCGAGATAGGAACATACAAATATAATTATTTACCATATGGGGTCAGTTCAGGACCTGGTGCATTTCAGCGTCTAATGACTAAAAAGTTGGAAGGGTCGCGGCGCGGCGCGGCGGGCGTGTGCGTGCGGGCGCGGCGCGGCGGCGGGCGGGCGGCGCTGCTGCGGCGGCTGGCTCGCTGGCCGCGCCTGCACCACGACCGCCTCTCCTTCACCGTCGACGTCTTGTACCACTCGGACGAAGCCACGATCTTCAGAGTGGGCGGCGTCGTCAAATTGACGTCTGATTGTCTTGCTGACGAAGAAGAGATGATCTACTTCAGCAGGACCTTGGTGCTGAAGGCTGAAGGCACGACAGAATACAAGATACATAACGAAATGCTTTACTGGGACGAAGTCACCGACCAAACTGCAAAAGCCGCCTTCCAGGTAACGGAGGCATACAATTGA
Protein
MSLISNLKSFDGEKWSVFQEQLDCFFIVNEVSDEKKVPLLITKLAPKVYEVLSYLCKPKKPQDLKFDQLCFKLAAKYDQKLPSAIERANFRQRNQAPNEKVEDYVLELRKLAAKCNFKDMEDQVKEKLMDGVSSKMIKFELMKSAGDATLERCIELARTVEAAILQTNGATNEVAEIFVSKSTNKSPQQQRNTYQQKNQCYCCGKWNHLKRDCRLNRKYCSECGIQGHIFRMCPKKQRKVNVLQEDNQQKDKNTDNEPTEFEPKYYETYACNTVSKIPPYYITIKMEECKLPFQLDTGSEVTVITENDKNDILINHVLQDCDIIFKNFDQSTTRPLGIVHNISVEYGNRSVYLKAFVVKNKSPRIIGRDWLNALDAQNSIEKKFAAVFSPGWGNFKGDVINLVLKKEVEPKCLPVRRVPYALKDKVNNEIKRLLDNGRITVVKHSKWGTPVVPILKSDGSVRLCGDYKLTVNKCLEVDHFPLPHVEDIINTLQGGEYYCELDLKEAYLQAPLSLESQDCTTIVTEIGTYKYNYLPYGVSSGPGAFQRLMTKKLEGSRRGAAGVCVRARRGGGRAALLRRLARWPRLHHDRLSFTVDVLYHSDEATIFRVGGVVKLTSDCLADEEEMIYFSRTLVLKAEGTTEYKIHNEMLYWDEVTDQTAKAAFQVTEAYN
Summary
Uniprot
A0A3P8NHI5
A0A3P8QD94
A0A1B6DD07
A0A1B6CPB9
A0A1B6E6W0
A0A3Q1EQC9
+ More
A0A1A8C840 A0A2S2PU23 A0A1X7TIE8 A0A1B6CK52 A0A3P8RGT3 A0A3R7GU59 A0A1A8FCH6 A0A3S0ZSV8 A0A131XQT5 A0A0J7KL04 A0A3P8Q506 A0A1E1XG22 A0A3P8NG55 A0A1X7TH22 A0A1Y1LPV1 A0A3B3H2N7 A0A2I4C6V0 X1XT86 A0A085NRB3 A0A1X7TR37 J9KFN3 A0A3B3R045 A0A267FRX0 J9KZ86 A0A3P8NEQ7 A0A085M8Q0 A0A085LTS7 X1WXW3 A0A1Y1KCE7 J9LDN7 A0A1Y1KIG3 A0A0V1LU74 A0A0V0TT39 A0A267GH18 A0A267EVG4 A2TGR4 J9KQR5 A0A3S2PB62 A0A267G4Q7 A0A0J7KF98 A2TGR5 A0A2R5LIR9 A0A1B8XWC1 A0A1S3J391 J9L709 A0A267F699 A0A1Y1NFA4 A0A1Y1MAK2 A0A3R7FFB4 A0A0J7KBM1 J9L142 X1WTX7 A0A2G8L891 A0A1Y1LR51 A0A2S2PLE8 A0A224Z2A6 A0A267F772 A0A2R2MK13 A0A267G5M8 A0A267GNB6 A0A293MR82 A0A3P8VSE7 K7JW34 A0A085MRV2 A0A085LME0 A0A1Y1LSF9 A0A1Y1LTC6 A0A085N4P6 A0A3S1BK89 A0A0V0RGX3 A0A085LV84 A0A0V1M0M6 A0A1Y1N2I4 A0A131Z1D5 A0A0A9WT47 A0A3P8VLQ9 A0A1W7R676 A0A0V0USX8 A0A1W7R6B8 A0A085LQ67 A0A085ND74 A0A0V1KNI7 A0A085LR60 A0A0V0RTT5 A0A2S2N957 A0A0V1M2E2 A0A085NBL6 A0A085M9N6 A0A0V0VH28 A0A0V1BVI1 A0A085MU81
A0A1A8C840 A0A2S2PU23 A0A1X7TIE8 A0A1B6CK52 A0A3P8RGT3 A0A3R7GU59 A0A1A8FCH6 A0A3S0ZSV8 A0A131XQT5 A0A0J7KL04 A0A3P8Q506 A0A1E1XG22 A0A3P8NG55 A0A1X7TH22 A0A1Y1LPV1 A0A3B3H2N7 A0A2I4C6V0 X1XT86 A0A085NRB3 A0A1X7TR37 J9KFN3 A0A3B3R045 A0A267FRX0 J9KZ86 A0A3P8NEQ7 A0A085M8Q0 A0A085LTS7 X1WXW3 A0A1Y1KCE7 J9LDN7 A0A1Y1KIG3 A0A0V1LU74 A0A0V0TT39 A0A267GH18 A0A267EVG4 A2TGR4 J9KQR5 A0A3S2PB62 A0A267G4Q7 A0A0J7KF98 A2TGR5 A0A2R5LIR9 A0A1B8XWC1 A0A1S3J391 J9L709 A0A267F699 A0A1Y1NFA4 A0A1Y1MAK2 A0A3R7FFB4 A0A0J7KBM1 J9L142 X1WTX7 A0A2G8L891 A0A1Y1LR51 A0A2S2PLE8 A0A224Z2A6 A0A267F772 A0A2R2MK13 A0A267G5M8 A0A267GNB6 A0A293MR82 A0A3P8VSE7 K7JW34 A0A085MRV2 A0A085LME0 A0A1Y1LSF9 A0A1Y1LTC6 A0A085N4P6 A0A3S1BK89 A0A0V0RGX3 A0A085LV84 A0A0V1M0M6 A0A1Y1N2I4 A0A131Z1D5 A0A0A9WT47 A0A3P8VLQ9 A0A1W7R676 A0A0V0USX8 A0A1W7R6B8 A0A085LQ67 A0A085ND74 A0A0V1KNI7 A0A085LR60 A0A0V0RTT5 A0A2S2N957 A0A0V1M2E2 A0A085NBL6 A0A085M9N6 A0A0V0VH28 A0A0V1BVI1 A0A085MU81
Pubmed
EMBL
GEDC01013717
JAS23581.1
GEDC01021971
JAS15327.1
GEDC01007675
GEDC01003661
+ More
JAS29623.1 JAS33637.1 HADZ01011823 HAEA01016148 SBP75764.1 GGMR01020219 MBY32838.1 GEDC01023506 GEDC01023205 JAS13792.1 JAS14093.1 NIRI01002335 RJW62347.1 HAEB01010230 SBQ56757.1 RQTK01000185 RUS85038.1 GEFM01006397 JAP69399.1 LBMM01006119 KMQ90911.1 GFAC01001149 JAT98039.1 GEZM01051399 JAV74971.1 ABLF02012266 ABLF02012271 KL367479 KFD72009.1 ABLF02036937 NIVC01000860 PAA75827.1 ABLF02026830 ABLF02041879 KL363215 KFD53596.1 KL363295 KFD48373.1 ABLF02006499 GEZM01092768 GEZM01092767 JAV56467.1 ABLF02030002 GEZM01085308 JAV60010.1 JYDW01000004 KRZ62956.1 JYDJ01000151 KRX42156.1 NIVC01000363 PAA84717.1 NIVC01001652 PAA65446.1 EF199621 ABM90392.1 RSAL01000130 RVE46453.1 NIVC01000555 PAA80976.1 LBMM01008360 KMQ88942.1 EF199622 ABM90393.1 GGLE01005288 MBY09414.1 KV461060 OCA14951.1 ABLF02016687 ABLF02016690 NIVC01001377 PAA68677.1 GEZM01009753 JAV94337.1 GEZM01036245 JAV82862.1 NIRI01001696 RJW65585.1 LBMM01010146 KMQ87646.1 ABLF02012827 ABLF02012828 ABLF02015552 ABLF02041322 ABLF02008248 ABLF02008249 ABLF02008250 ABLF02035153 ABLF02037855 ABLF02037866 ABLF02061872 MRZV01000174 PIK56483.1 GEZM01050895 JAV75318.1 GGMR01017626 MBY30245.1 GFPF01012842 MAA23988.1 NIVC01001308 PAA69606.1 NIVC01000538 PAA81343.1 NIVC01000264 PAA86792.1 GFWV01017841 MAA42569.1 AAZX01023237 KL367711 KFD59948.1 KL363391 KFD46136.1 GEZM01051546 GEZM01051543 JAV74825.1 GEZM01051545 GEZM01051542 JAV74826.1 KL367556 KFD64442.1 RQTK01001807 RUS69146.1 JYDL01000185 KRX13745.1 KL363282 KFD48880.1 JYDO01000426 KRZ65264.1 GEZM01014356 JAV92133.1 GEDV01003323 JAP85234.1 GBHO01031997 JAG11607.1 GEHC01000976 JAV46669.1 JYDN01000181 KRX54109.1 GEHC01000975 JAV46670.1 KL363340 KFD47113.1 KL367515 KFD67420.1 JYDW01000348 KRZ48953.1 KL363326 KFD47456.1 JYDL01000080 KRX17894.1 GGMR01001051 MBY13670.1 JYDO01000284 KRZ65891.1 KL367520 KFD66862.1 KL363212 KFD53932.1 JYDN01000036 KRX62850.1 JYDH01000010 KRY40903.1 KL367649 KFD60777.1
JAS29623.1 JAS33637.1 HADZ01011823 HAEA01016148 SBP75764.1 GGMR01020219 MBY32838.1 GEDC01023506 GEDC01023205 JAS13792.1 JAS14093.1 NIRI01002335 RJW62347.1 HAEB01010230 SBQ56757.1 RQTK01000185 RUS85038.1 GEFM01006397 JAP69399.1 LBMM01006119 KMQ90911.1 GFAC01001149 JAT98039.1 GEZM01051399 JAV74971.1 ABLF02012266 ABLF02012271 KL367479 KFD72009.1 ABLF02036937 NIVC01000860 PAA75827.1 ABLF02026830 ABLF02041879 KL363215 KFD53596.1 KL363295 KFD48373.1 ABLF02006499 GEZM01092768 GEZM01092767 JAV56467.1 ABLF02030002 GEZM01085308 JAV60010.1 JYDW01000004 KRZ62956.1 JYDJ01000151 KRX42156.1 NIVC01000363 PAA84717.1 NIVC01001652 PAA65446.1 EF199621 ABM90392.1 RSAL01000130 RVE46453.1 NIVC01000555 PAA80976.1 LBMM01008360 KMQ88942.1 EF199622 ABM90393.1 GGLE01005288 MBY09414.1 KV461060 OCA14951.1 ABLF02016687 ABLF02016690 NIVC01001377 PAA68677.1 GEZM01009753 JAV94337.1 GEZM01036245 JAV82862.1 NIRI01001696 RJW65585.1 LBMM01010146 KMQ87646.1 ABLF02012827 ABLF02012828 ABLF02015552 ABLF02041322 ABLF02008248 ABLF02008249 ABLF02008250 ABLF02035153 ABLF02037855 ABLF02037866 ABLF02061872 MRZV01000174 PIK56483.1 GEZM01050895 JAV75318.1 GGMR01017626 MBY30245.1 GFPF01012842 MAA23988.1 NIVC01001308 PAA69606.1 NIVC01000538 PAA81343.1 NIVC01000264 PAA86792.1 GFWV01017841 MAA42569.1 AAZX01023237 KL367711 KFD59948.1 KL363391 KFD46136.1 GEZM01051546 GEZM01051543 JAV74825.1 GEZM01051545 GEZM01051542 JAV74826.1 KL367556 KFD64442.1 RQTK01001807 RUS69146.1 JYDL01000185 KRX13745.1 KL363282 KFD48880.1 JYDO01000426 KRZ65264.1 GEZM01014356 JAV92133.1 GEDV01003323 JAP85234.1 GBHO01031997 JAG11607.1 GEHC01000976 JAV46669.1 JYDN01000181 KRX54109.1 GEHC01000975 JAV46670.1 KL363340 KFD47113.1 KL367515 KFD67420.1 JYDW01000348 KRZ48953.1 KL363326 KFD47456.1 JYDL01000080 KRX17894.1 GGMR01001051 MBY13670.1 JYDO01000284 KRZ65891.1 KL367520 KFD66862.1 KL363212 KFD53932.1 JYDN01000036 KRX62850.1 JYDH01000010 KRY40903.1 KL367649 KFD60777.1
Proteomes
Pfam
Interpro
IPR021109
Peptidase_aspartic_dom_sf
+ More
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR000477 RT_dom
IPR001584 Integrase_cat-core
IPR041577 RT_RNaseH_2
IPR012337 RNaseH-like_sf
IPR036875 Znf_CCHC_sf
IPR001878 Znf_CCHC
IPR001995 Peptidase_A2_cat
IPR001969 Aspartic_peptidase_AS
IPR001753 Enoyl-CoA_hydra/iso
IPR029045 ClpP/crotonase-like_dom_sf
IPR041373 RT_RNaseH
IPR005162 Retrotrans_gag_dom
IPR016024 ARM-type_fold
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR008383 API5
IPR038718 SNF2-like_sf
IPR027417 P-loop_NTPase
IPR000330 SNF2_N
IPR034128 K02A2.6-like
IPR006612 THAP_Znf
IPR018061 Retropepsins
IPR009068 S15_NS1_RNA-bd
IPR013103 RVT_2
IPR025724 GAG-pre-integrase_dom
IPR008936 Rho_GTPase_activation_prot
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR000477 RT_dom
IPR001584 Integrase_cat-core
IPR041577 RT_RNaseH_2
IPR012337 RNaseH-like_sf
IPR036875 Znf_CCHC_sf
IPR001878 Znf_CCHC
IPR001995 Peptidase_A2_cat
IPR001969 Aspartic_peptidase_AS
IPR001753 Enoyl-CoA_hydra/iso
IPR029045 ClpP/crotonase-like_dom_sf
IPR041373 RT_RNaseH
IPR005162 Retrotrans_gag_dom
IPR016024 ARM-type_fold
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR008383 API5
IPR038718 SNF2-like_sf
IPR027417 P-loop_NTPase
IPR000330 SNF2_N
IPR034128 K02A2.6-like
IPR006612 THAP_Znf
IPR018061 Retropepsins
IPR009068 S15_NS1_RNA-bd
IPR013103 RVT_2
IPR025724 GAG-pre-integrase_dom
IPR008936 Rho_GTPase_activation_prot
SUPFAM
Gene 3D
ProteinModelPortal
A0A3P8NHI5
A0A3P8QD94
A0A1B6DD07
A0A1B6CPB9
A0A1B6E6W0
A0A3Q1EQC9
+ More
A0A1A8C840 A0A2S2PU23 A0A1X7TIE8 A0A1B6CK52 A0A3P8RGT3 A0A3R7GU59 A0A1A8FCH6 A0A3S0ZSV8 A0A131XQT5 A0A0J7KL04 A0A3P8Q506 A0A1E1XG22 A0A3P8NG55 A0A1X7TH22 A0A1Y1LPV1 A0A3B3H2N7 A0A2I4C6V0 X1XT86 A0A085NRB3 A0A1X7TR37 J9KFN3 A0A3B3R045 A0A267FRX0 J9KZ86 A0A3P8NEQ7 A0A085M8Q0 A0A085LTS7 X1WXW3 A0A1Y1KCE7 J9LDN7 A0A1Y1KIG3 A0A0V1LU74 A0A0V0TT39 A0A267GH18 A0A267EVG4 A2TGR4 J9KQR5 A0A3S2PB62 A0A267G4Q7 A0A0J7KF98 A2TGR5 A0A2R5LIR9 A0A1B8XWC1 A0A1S3J391 J9L709 A0A267F699 A0A1Y1NFA4 A0A1Y1MAK2 A0A3R7FFB4 A0A0J7KBM1 J9L142 X1WTX7 A0A2G8L891 A0A1Y1LR51 A0A2S2PLE8 A0A224Z2A6 A0A267F772 A0A2R2MK13 A0A267G5M8 A0A267GNB6 A0A293MR82 A0A3P8VSE7 K7JW34 A0A085MRV2 A0A085LME0 A0A1Y1LSF9 A0A1Y1LTC6 A0A085N4P6 A0A3S1BK89 A0A0V0RGX3 A0A085LV84 A0A0V1M0M6 A0A1Y1N2I4 A0A131Z1D5 A0A0A9WT47 A0A3P8VLQ9 A0A1W7R676 A0A0V0USX8 A0A1W7R6B8 A0A085LQ67 A0A085ND74 A0A0V1KNI7 A0A085LR60 A0A0V0RTT5 A0A2S2N957 A0A0V1M2E2 A0A085NBL6 A0A085M9N6 A0A0V0VH28 A0A0V1BVI1 A0A085MU81
A0A1A8C840 A0A2S2PU23 A0A1X7TIE8 A0A1B6CK52 A0A3P8RGT3 A0A3R7GU59 A0A1A8FCH6 A0A3S0ZSV8 A0A131XQT5 A0A0J7KL04 A0A3P8Q506 A0A1E1XG22 A0A3P8NG55 A0A1X7TH22 A0A1Y1LPV1 A0A3B3H2N7 A0A2I4C6V0 X1XT86 A0A085NRB3 A0A1X7TR37 J9KFN3 A0A3B3R045 A0A267FRX0 J9KZ86 A0A3P8NEQ7 A0A085M8Q0 A0A085LTS7 X1WXW3 A0A1Y1KCE7 J9LDN7 A0A1Y1KIG3 A0A0V1LU74 A0A0V0TT39 A0A267GH18 A0A267EVG4 A2TGR4 J9KQR5 A0A3S2PB62 A0A267G4Q7 A0A0J7KF98 A2TGR5 A0A2R5LIR9 A0A1B8XWC1 A0A1S3J391 J9L709 A0A267F699 A0A1Y1NFA4 A0A1Y1MAK2 A0A3R7FFB4 A0A0J7KBM1 J9L142 X1WTX7 A0A2G8L891 A0A1Y1LR51 A0A2S2PLE8 A0A224Z2A6 A0A267F772 A0A2R2MK13 A0A267G5M8 A0A267GNB6 A0A293MR82 A0A3P8VSE7 K7JW34 A0A085MRV2 A0A085LME0 A0A1Y1LSF9 A0A1Y1LTC6 A0A085N4P6 A0A3S1BK89 A0A0V0RGX3 A0A085LV84 A0A0V1M0M6 A0A1Y1N2I4 A0A131Z1D5 A0A0A9WT47 A0A3P8VLQ9 A0A1W7R676 A0A0V0USX8 A0A1W7R6B8 A0A085LQ67 A0A085ND74 A0A0V1KNI7 A0A085LR60 A0A0V0RTT5 A0A2S2N957 A0A0V1M2E2 A0A085NBL6 A0A085M9N6 A0A0V0VH28 A0A0V1BVI1 A0A085MU81
PDB
4OL8
E-value=2.46979e-18,
Score=228
Ontologies
PATHWAY
GO
Topology
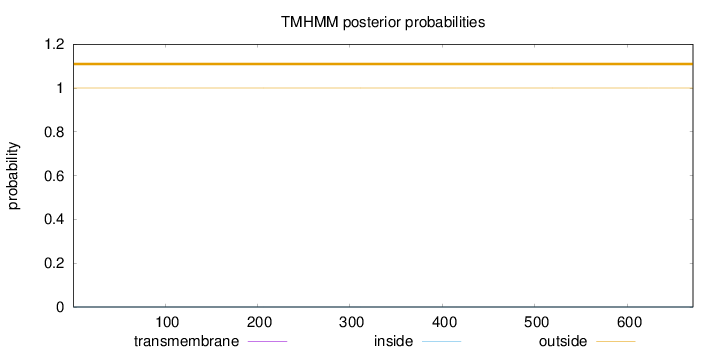
Length:
671
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00022
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00008
outside
1 - 671
Population Genetic Test Statistics
Pi
4.333924
Theta
6.889869
Tajima's D
-1.364791
CLR
109.099853
CSRT
0.0736463176841158
Interpretation
Uncertain
