Gene
KWMTBOMO11153
Pre Gene Modal
BGIBMGA008584
Annotation
Rh-like_protein_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.797
Sequence
CDS
ATGATTAAGTACACTGCCTTCAAGTACGCCCCGGTGTTGGCATTTCAACTACTTCTGATGGTGCTGTATTTTATATTCGTGAGATACGGAGGCGGTGAGGATGTTAATATTAAAGGGTTCGAAGATACTCATGTGATGATCTTCATCGGTTTCGGCTTTTTAATGACATTCCTCAAAAAGTACAGCTACAGCGCACTTGGGTTCAACTGGCTGTTGGCAGCCCTAGTCATTCAATGGGCACTACTCTGCCAGTCCTTCTACCACATGAAGAATAACACTATTTATGTCACTAAGAAGTCTTTGCTTGAAGCGGATATTATGTCTGCGACAGTTCTCATAACTTTTGGTGCACTATTAGGAGTAGCTAGTGGACTGCAGTTGTTGTTCATAGCTATTGTTGAAACGGCCGTTGCTTGCCTCAATATGTGGCTTGTGGCGGATGTATTTAAGGCATCTGATGTTGGTGGCTCGATAGCCATTCATACTTTTGGAGCATATTTTGGGATCGGTGTCAGCATGGCACTGAAATCAAAGAATAAGGACACAACTCCTACTACAAATGGAAACACGACTCCAAGCGTCGACCTGAATGCCCCTAGCTACATATCCGATGTCACTGCTATGATAGGATCAATATTTTTGTGGATATATTGGCCATCATTTAATTCTGGACTAACGAACACAGACGCCGAATATCAAAGAGCTGTTGTCAATACGTATTTATCTCTGGCTGCTGCTACGGTAACTACATTTGTTTTATCATCAGCACTGAATGAGCACAAAGGTAAATTTGACATGGTCCACATCCAGAACTCGACCCTGGCTGGTGGAGTGGCTGTGGGCTCCGTGTGTAACATGCAGATCGGAGCTGGTGGAGCAGTGGCTATTGGAATCGGCGCGGGGATCTTGAGCGTTCTAGGATATCGTTACCTGACACCTAAACTCACAAAAATTGGCATCCTGGACACTTGCGGTGTGAACAACCTTCACGGTATGCCTGGAATCTACTCCGGTGTTCTCTCCGTCCTGTTCGCCGGTCTAGCCACCAGCGACGCTTATGGCGCTGAACTTGGCACTGTTTTTAGTGGCATTGGCCAGAATGGCAGAACTATTGGACAACAAGCTTTGAACCAGTTCCTAGCTCTTGTGGTGACTCTGGTATTATCACTGGTATCTGGATTTCTAACAGGTCTCCTCTCAAAGATTCCAGCACTGGAGCCCATGAAGCAGAAAGAATGGTACAACGACGAAATCAACTGGGAATTACCTTAA
Protein
MIKYTAFKYAPVLAFQLLLMVLYFIFVRYGGGEDVNIKGFEDTHVMIFIGFGFLMTFLKKYSYSALGFNWLLAALVIQWALLCQSFYHMKNNTIYVTKKSLLEADIMSATVLITFGALLGVASGLQLLFIAIVETAVACLNMWLVADVFKASDVGGSIAIHTFGAYFGIGVSMALKSKNKDTTPTTNGNTTPSVDLNAPSYISDVTAMIGSIFLWIYWPSFNSGLTNTDAEYQRAVVNTYLSLAAATVTTFVLSSALNEHKGKFDMVHIQNSTLAGGVAVGSVCNMQIGAGGAVAIGIGAGILSVLGYRYLTPKLTKIGILDTCGVNNLHGMPGIYSGVLSVLFAGLATSDAYGAELGTVFSGIGQNGRTIGQQALNQFLALVVTLVLSLVSGFLTGLLSKIPALEPMKQKEWYNDEINWELP
Summary
Similarity
Belongs to the ammonium transporter (TC 2.A.49) family. Rh subfamily.
Uniprot
Q1L6Q3
H9JGD7
Q0GGW0
A0A3S2PBS4
A0A212FFQ0
A0A2A4J531
+ More
A0A194QI59 A0A182H440 D0V1X1 A0A182H441 A0A0A1XTK6 B0WXN8 A0A0K8VX06 W8BHM7 A0A1Q3FSC6 Q5BP95 A0A182NCF3 A0A034VYH8 A0A1B0B0C1 A0A182XVX2 A0A0L0BMH4 A0A1W7R848 A0A1Y1K678 A0A1A9WLE9 A0A1A9VAL3 Q16ZV6 A0A182VZ15 Q5BP96 A0A182XBB3 A0A182KLQ8 A0A182TXI9 Q7Z1M3 A0A182V8S9 A0A182R6C7 A0A182I056 A0A182QR18 A0A1Y1K5H6 A0A182PKK8 A0A182FKT0 A0A1J1IJE5 W5JFW6 A0A182K9T4 A0A1I8MNV0 A0A182LUJ8 A0A1B0GIJ7 T1PDH0 A0A0P5YIS2 A0A182SKD0 A0A182J9G1 A0A0P5RPQ0 A0A0P5NX88 A0A0P5NXQ8 A0A0P6D083 A0A0P6H0A2 A0A0P6EBJ5 A0A0P5LTM1 A0A0P5HZN3 A0A0P5MMZ8 A0A0P5PU14 A0A0P6GFI7 A0A0P6EDN5 A0A0P5X208 A0A0P5KM51 A0A0P6AFM8 A0A1B0D511 A0A1Y0AWM4 A0A0P5YXY5 A0A0P5WVX6 A0A084VHD3 A0A0P4ZGM0 A0A0P5UYC7 A0A0P9AJ02 A0A0P5EEU7 B3M9L6 A0A0P5PE09 A0A0P5GG19 B4PIC8 B4KXC7 Q9V3T3 A0A0R1DZ65 A0A2P8XZM1 Q4VUH9 Q1L6Q2 A0A1W4VN70 M9PEN8 A0A0R4J8N2 A0A0P5VAR8 A0A3B0K2D2 E9HAS9 A0A0J9RPN4 A0A3B0J6E3 A0A0P5V4P5 A0A0P5REI6 B3NG78 B4HUC1 A0A0P5LJX2 A0A0P6HKJ2 A0A0P5LU76 A0A0P6F331 A0A0P5LAL5
A0A194QI59 A0A182H440 D0V1X1 A0A182H441 A0A0A1XTK6 B0WXN8 A0A0K8VX06 W8BHM7 A0A1Q3FSC6 Q5BP95 A0A182NCF3 A0A034VYH8 A0A1B0B0C1 A0A182XVX2 A0A0L0BMH4 A0A1W7R848 A0A1Y1K678 A0A1A9WLE9 A0A1A9VAL3 Q16ZV6 A0A182VZ15 Q5BP96 A0A182XBB3 A0A182KLQ8 A0A182TXI9 Q7Z1M3 A0A182V8S9 A0A182R6C7 A0A182I056 A0A182QR18 A0A1Y1K5H6 A0A182PKK8 A0A182FKT0 A0A1J1IJE5 W5JFW6 A0A182K9T4 A0A1I8MNV0 A0A182LUJ8 A0A1B0GIJ7 T1PDH0 A0A0P5YIS2 A0A182SKD0 A0A182J9G1 A0A0P5RPQ0 A0A0P5NX88 A0A0P5NXQ8 A0A0P6D083 A0A0P6H0A2 A0A0P6EBJ5 A0A0P5LTM1 A0A0P5HZN3 A0A0P5MMZ8 A0A0P5PU14 A0A0P6GFI7 A0A0P6EDN5 A0A0P5X208 A0A0P5KM51 A0A0P6AFM8 A0A1B0D511 A0A1Y0AWM4 A0A0P5YXY5 A0A0P5WVX6 A0A084VHD3 A0A0P4ZGM0 A0A0P5UYC7 A0A0P9AJ02 A0A0P5EEU7 B3M9L6 A0A0P5PE09 A0A0P5GG19 B4PIC8 B4KXC7 Q9V3T3 A0A0R1DZ65 A0A2P8XZM1 Q4VUH9 Q1L6Q2 A0A1W4VN70 M9PEN8 A0A0R4J8N2 A0A0P5VAR8 A0A3B0K2D2 E9HAS9 A0A0J9RPN4 A0A3B0J6E3 A0A0P5V4P5 A0A0P5REI6 B3NG78 B4HUC1 A0A0P5LJX2 A0A0P6HKJ2 A0A0P5LU76 A0A0P6F331 A0A0P5LAL5
Pubmed
19121390
22118469
26354079
26483478
20561978
25830018
+ More
24495485 25348373 25244985 26108605 28004739 17510324 20966253 12364791 14747013 16227429 17210077 20920257 23761445 25315136 28341416 24438588 17994087 17550304 10852913 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29403074 15632085 21292972 22936249
24495485 25348373 25244985 26108605 28004739 17510324 20966253 12364791 14747013 16227429 17210077 20920257 23761445 25315136 28341416 24438588 17994087 17550304 10852913 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29403074 15632085 21292972 22936249
EMBL
DQ013063
AAY81951.1
BABH01033582
DQ864985
ABI20766.1
RSAL01000116
+ More
RVE46843.1 AGBW02008787 OWR52557.1 NWSH01003203 PCG66788.1 KQ458880 KPJ04615.1 JXUM01108705 KQ565257 KXJ71166.1 GU013469 ACX71870.1 JXUM01108707 JXUM01108708 KXJ71167.1 GBXI01004920 GBXI01000079 JAD09372.1 JAD14213.1 DS232169 EDS36568.1 GDHF01009219 JAI43095.1 GAMC01013804 JAB92751.1 GFDL01004580 JAV30465.1 AY926464 AAX19514.1 GAKP01011469 GAKP01011468 JAC47484.1 JXJN01006701 JXJN01006702 JXJN01006703 JXJN01006704 JXJN01006705 JRES01001653 KNC21133.1 GEHC01000362 JAV47283.1 GEZM01094318 JAV55710.1 CH477482 EAT40220.1 AY926463 AAX19513.1 AF510715 AAAB01008987 AAP47144.1 EAA01247.4 APCN01002045 AXCN02000019 GEZM01094317 JAV55711.1 CVRI01000049 CRK99182.1 ADMH02001343 ETN62951.1 AXCM01002551 AJWK01014215 KA646175 AFP60804.1 GDIP01058716 JAM44999.1 GDIQ01222495 GDIQ01116588 GDIQ01116587 JAL35138.1 GDIQ01140896 JAL10830.1 GDIQ01229940 GDIQ01138840 GDIQ01136288 GDIQ01136287 GDIQ01123980 GDIQ01121530 GDIP01078748 GDIQ01073304 GDIQ01073303 JAL15439.1 JAM24967.1 GDIQ01084897 JAN09840.1 GDIQ01179460 GDIQ01179459 GDIQ01026561 JAN68176.1 GDIQ01065113 GDIQ01065112 JAN29625.1 GDIQ01165330 JAK86395.1 GDIQ01251053 GDIQ01251000 GDIQ01233546 GDIQ01233545 GDIQ01229939 GDIQ01220808 GDIQ01220806 GDIQ01197154 GDIQ01195458 GDIQ01161178 GDIQ01161177 GDIQ01159620 GDIQ01138839 GDIQ01136286 GDIQ01127438 GDIQ01127437 GDIQ01123979 GDIQ01121529 GDIQ01109286 GDIQ01097289 GDIP01093346 GDIQ01086547 GDIQ01081956 GDIQ01081955 GDIQ01073302 GDIQ01051063 GDIQ01051061 LRGB01000359 JAK30917.1 JAM10369.1 KZS19565.1 GDIQ01165331 GDIQ01165329 JAK86396.1 GDIQ01125784 JAL25942.1 GDIQ01033926 JAN60811.1 GDIQ01253706 GDIQ01251001 GDIQ01220807 GDIQ01198564 GDIQ01161179 GDIQ01159621 GDIQ01128793 GDIQ01127436 GDIQ01125785 GDIQ01098748 GDIQ01095537 GDIQ01091140 GDIQ01078449 GDIQ01051062 GDIQ01046769 GDIQ01044623 GDIQ01030528 GDIQ01029940 JAN16288.1 GDIP01078749 JAM24966.1 GDIQ01183316 GDIQ01183315 JAK68409.1 GDIP01031549 JAM72166.1 AJVK01011685 AJVK01011686 KY921808 ART29397.1 GDIP01064960 JAM38755.1 GDIP01081296 JAM22419.1 ATLV01013152 KE524842 KFB37377.1 GDIP01213092 JAJ10310.1 GDIP01108395 JAL95319.1 CH902618 KPU77817.1 GDIP01143951 JAJ79451.1 KZS19566.1 EDV39022.2 GDIQ01129985 JAL21741.1 GDIQ01247302 JAK04423.1 CM000159 EDW93470.1 CH933809 EDW17585.1 AF181624 AF172639 AF193812 AE014296 AH009249 AF218885 AF218887 AAD55410.1 AAF09188.1 AAF19374.1 AAF47951.1 AAF64673.1 KRK01219.1 PYGN01001118 PSN37449.1 AY622225 AAV40852.1 DQ013064 CM000363 AAY81952.1 EDX09276.1 AGB94129.1 AHN57963.1 CH379070 EAL30167.2 GDIP01102694 JAM01021.1 OUUW01000002 SPP77548.1 GL732613 EFX71083.1 CM002912 KMY97692.1 SPP77547.1 GDIP01105552 JAL98162.1 GDIQ01111231 JAL40495.1 CH954178 EDV50979.2 CH480817 EDW50542.1 GDIQ01169876 JAK81849.1 GDIQ01031871 JAN62866.1 GDIQ01173909 JAK77816.1 GDIQ01176207 GDIQ01066015 GDIQ01066014 GDIQ01057500 JAN28722.1 GDIQ01173908 JAK77817.1
RVE46843.1 AGBW02008787 OWR52557.1 NWSH01003203 PCG66788.1 KQ458880 KPJ04615.1 JXUM01108705 KQ565257 KXJ71166.1 GU013469 ACX71870.1 JXUM01108707 JXUM01108708 KXJ71167.1 GBXI01004920 GBXI01000079 JAD09372.1 JAD14213.1 DS232169 EDS36568.1 GDHF01009219 JAI43095.1 GAMC01013804 JAB92751.1 GFDL01004580 JAV30465.1 AY926464 AAX19514.1 GAKP01011469 GAKP01011468 JAC47484.1 JXJN01006701 JXJN01006702 JXJN01006703 JXJN01006704 JXJN01006705 JRES01001653 KNC21133.1 GEHC01000362 JAV47283.1 GEZM01094318 JAV55710.1 CH477482 EAT40220.1 AY926463 AAX19513.1 AF510715 AAAB01008987 AAP47144.1 EAA01247.4 APCN01002045 AXCN02000019 GEZM01094317 JAV55711.1 CVRI01000049 CRK99182.1 ADMH02001343 ETN62951.1 AXCM01002551 AJWK01014215 KA646175 AFP60804.1 GDIP01058716 JAM44999.1 GDIQ01222495 GDIQ01116588 GDIQ01116587 JAL35138.1 GDIQ01140896 JAL10830.1 GDIQ01229940 GDIQ01138840 GDIQ01136288 GDIQ01136287 GDIQ01123980 GDIQ01121530 GDIP01078748 GDIQ01073304 GDIQ01073303 JAL15439.1 JAM24967.1 GDIQ01084897 JAN09840.1 GDIQ01179460 GDIQ01179459 GDIQ01026561 JAN68176.1 GDIQ01065113 GDIQ01065112 JAN29625.1 GDIQ01165330 JAK86395.1 GDIQ01251053 GDIQ01251000 GDIQ01233546 GDIQ01233545 GDIQ01229939 GDIQ01220808 GDIQ01220806 GDIQ01197154 GDIQ01195458 GDIQ01161178 GDIQ01161177 GDIQ01159620 GDIQ01138839 GDIQ01136286 GDIQ01127438 GDIQ01127437 GDIQ01123979 GDIQ01121529 GDIQ01109286 GDIQ01097289 GDIP01093346 GDIQ01086547 GDIQ01081956 GDIQ01081955 GDIQ01073302 GDIQ01051063 GDIQ01051061 LRGB01000359 JAK30917.1 JAM10369.1 KZS19565.1 GDIQ01165331 GDIQ01165329 JAK86396.1 GDIQ01125784 JAL25942.1 GDIQ01033926 JAN60811.1 GDIQ01253706 GDIQ01251001 GDIQ01220807 GDIQ01198564 GDIQ01161179 GDIQ01159621 GDIQ01128793 GDIQ01127436 GDIQ01125785 GDIQ01098748 GDIQ01095537 GDIQ01091140 GDIQ01078449 GDIQ01051062 GDIQ01046769 GDIQ01044623 GDIQ01030528 GDIQ01029940 JAN16288.1 GDIP01078749 JAM24966.1 GDIQ01183316 GDIQ01183315 JAK68409.1 GDIP01031549 JAM72166.1 AJVK01011685 AJVK01011686 KY921808 ART29397.1 GDIP01064960 JAM38755.1 GDIP01081296 JAM22419.1 ATLV01013152 KE524842 KFB37377.1 GDIP01213092 JAJ10310.1 GDIP01108395 JAL95319.1 CH902618 KPU77817.1 GDIP01143951 JAJ79451.1 KZS19566.1 EDV39022.2 GDIQ01129985 JAL21741.1 GDIQ01247302 JAK04423.1 CM000159 EDW93470.1 CH933809 EDW17585.1 AF181624 AF172639 AF193812 AE014296 AH009249 AF218885 AF218887 AAD55410.1 AAF09188.1 AAF19374.1 AAF47951.1 AAF64673.1 KRK01219.1 PYGN01001118 PSN37449.1 AY622225 AAV40852.1 DQ013064 CM000363 AAY81952.1 EDX09276.1 AGB94129.1 AHN57963.1 CH379070 EAL30167.2 GDIP01102694 JAM01021.1 OUUW01000002 SPP77548.1 GL732613 EFX71083.1 CM002912 KMY97692.1 SPP77547.1 GDIP01105552 JAL98162.1 GDIQ01111231 JAL40495.1 CH954178 EDV50979.2 CH480817 EDW50542.1 GDIQ01169876 JAK81849.1 GDIQ01031871 JAN62866.1 GDIQ01173909 JAK77816.1 GDIQ01176207 GDIQ01066015 GDIQ01066014 GDIQ01057500 JAN28722.1 GDIQ01173908 JAK77817.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000218220
UP000053268
UP000069940
+ More
UP000249989 UP000002320 UP000075884 UP000092460 UP000076408 UP000037069 UP000091820 UP000078200 UP000008820 UP000075920 UP000076407 UP000075882 UP000075902 UP000007062 UP000075903 UP000075900 UP000075840 UP000075886 UP000075885 UP000069272 UP000183832 UP000000673 UP000075881 UP000095301 UP000075883 UP000092461 UP000075901 UP000075880 UP000076858 UP000092462 UP000030765 UP000007801 UP000002282 UP000009192 UP000000803 UP000245037 UP000000304 UP000192221 UP000001819 UP000268350 UP000000305 UP000008711 UP000001292
UP000249989 UP000002320 UP000075884 UP000092460 UP000076408 UP000037069 UP000091820 UP000078200 UP000008820 UP000075920 UP000076407 UP000075882 UP000075902 UP000007062 UP000075903 UP000075900 UP000075840 UP000075886 UP000075885 UP000069272 UP000183832 UP000000673 UP000075881 UP000095301 UP000075883 UP000092461 UP000075901 UP000075880 UP000076858 UP000092462 UP000030765 UP000007801 UP000002282 UP000009192 UP000000803 UP000245037 UP000000304 UP000192221 UP000001819 UP000268350 UP000000305 UP000008711 UP000001292
Pfam
PF00909 Ammonium_transp
Interpro
Gene 3D
ProteinModelPortal
Q1L6Q3
H9JGD7
Q0GGW0
A0A3S2PBS4
A0A212FFQ0
A0A2A4J531
+ More
A0A194QI59 A0A182H440 D0V1X1 A0A182H441 A0A0A1XTK6 B0WXN8 A0A0K8VX06 W8BHM7 A0A1Q3FSC6 Q5BP95 A0A182NCF3 A0A034VYH8 A0A1B0B0C1 A0A182XVX2 A0A0L0BMH4 A0A1W7R848 A0A1Y1K678 A0A1A9WLE9 A0A1A9VAL3 Q16ZV6 A0A182VZ15 Q5BP96 A0A182XBB3 A0A182KLQ8 A0A182TXI9 Q7Z1M3 A0A182V8S9 A0A182R6C7 A0A182I056 A0A182QR18 A0A1Y1K5H6 A0A182PKK8 A0A182FKT0 A0A1J1IJE5 W5JFW6 A0A182K9T4 A0A1I8MNV0 A0A182LUJ8 A0A1B0GIJ7 T1PDH0 A0A0P5YIS2 A0A182SKD0 A0A182J9G1 A0A0P5RPQ0 A0A0P5NX88 A0A0P5NXQ8 A0A0P6D083 A0A0P6H0A2 A0A0P6EBJ5 A0A0P5LTM1 A0A0P5HZN3 A0A0P5MMZ8 A0A0P5PU14 A0A0P6GFI7 A0A0P6EDN5 A0A0P5X208 A0A0P5KM51 A0A0P6AFM8 A0A1B0D511 A0A1Y0AWM4 A0A0P5YXY5 A0A0P5WVX6 A0A084VHD3 A0A0P4ZGM0 A0A0P5UYC7 A0A0P9AJ02 A0A0P5EEU7 B3M9L6 A0A0P5PE09 A0A0P5GG19 B4PIC8 B4KXC7 Q9V3T3 A0A0R1DZ65 A0A2P8XZM1 Q4VUH9 Q1L6Q2 A0A1W4VN70 M9PEN8 A0A0R4J8N2 A0A0P5VAR8 A0A3B0K2D2 E9HAS9 A0A0J9RPN4 A0A3B0J6E3 A0A0P5V4P5 A0A0P5REI6 B3NG78 B4HUC1 A0A0P5LJX2 A0A0P6HKJ2 A0A0P5LU76 A0A0P6F331 A0A0P5LAL5
A0A194QI59 A0A182H440 D0V1X1 A0A182H441 A0A0A1XTK6 B0WXN8 A0A0K8VX06 W8BHM7 A0A1Q3FSC6 Q5BP95 A0A182NCF3 A0A034VYH8 A0A1B0B0C1 A0A182XVX2 A0A0L0BMH4 A0A1W7R848 A0A1Y1K678 A0A1A9WLE9 A0A1A9VAL3 Q16ZV6 A0A182VZ15 Q5BP96 A0A182XBB3 A0A182KLQ8 A0A182TXI9 Q7Z1M3 A0A182V8S9 A0A182R6C7 A0A182I056 A0A182QR18 A0A1Y1K5H6 A0A182PKK8 A0A182FKT0 A0A1J1IJE5 W5JFW6 A0A182K9T4 A0A1I8MNV0 A0A182LUJ8 A0A1B0GIJ7 T1PDH0 A0A0P5YIS2 A0A182SKD0 A0A182J9G1 A0A0P5RPQ0 A0A0P5NX88 A0A0P5NXQ8 A0A0P6D083 A0A0P6H0A2 A0A0P6EBJ5 A0A0P5LTM1 A0A0P5HZN3 A0A0P5MMZ8 A0A0P5PU14 A0A0P6GFI7 A0A0P6EDN5 A0A0P5X208 A0A0P5KM51 A0A0P6AFM8 A0A1B0D511 A0A1Y0AWM4 A0A0P5YXY5 A0A0P5WVX6 A0A084VHD3 A0A0P4ZGM0 A0A0P5UYC7 A0A0P9AJ02 A0A0P5EEU7 B3M9L6 A0A0P5PE09 A0A0P5GG19 B4PIC8 B4KXC7 Q9V3T3 A0A0R1DZ65 A0A2P8XZM1 Q4VUH9 Q1L6Q2 A0A1W4VN70 M9PEN8 A0A0R4J8N2 A0A0P5VAR8 A0A3B0K2D2 E9HAS9 A0A0J9RPN4 A0A3B0J6E3 A0A0P5V4P5 A0A0P5REI6 B3NG78 B4HUC1 A0A0P5LJX2 A0A0P6HKJ2 A0A0P5LU76 A0A0P6F331 A0A0P5LAL5
PDB
3HD6
E-value=9.0331e-70,
Score=670
Ontologies
GO
Topology
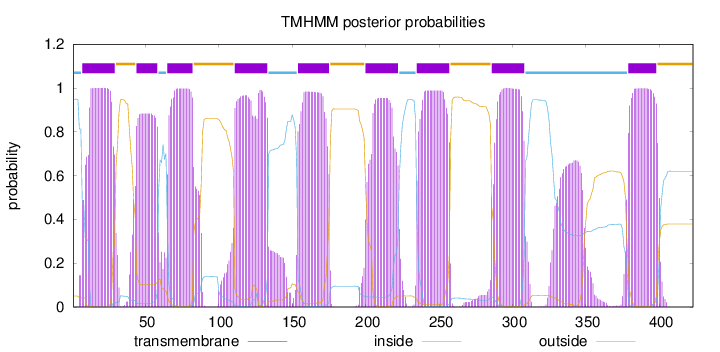
Length:
423
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
207.75646
Exp number, first 60 AAs:
36.66792
Total prob of N-in:
0.94927
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 43
TMhelix
44 - 58
inside
59 - 64
TMhelix
65 - 82
outside
83 - 110
TMhelix
111 - 133
inside
134 - 153
TMhelix
154 - 175
outside
176 - 199
TMhelix
200 - 222
inside
223 - 234
TMhelix
235 - 257
outside
258 - 285
TMhelix
286 - 308
inside
309 - 378
TMhelix
379 - 398
outside
399 - 423
Population Genetic Test Statistics
Pi
25.808612
Theta
17.743111
Tajima's D
-2.062867
CLR
2.310094
CSRT
0.00999950002499875
Interpretation
Possibly Positive selection
