Pre Gene Modal
BGIBMGA008579
Annotation
PREDICTED:_fatty_acid_synthase-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.464
Sequence
CDS
ATGGACTTCAGTGGCTATAATTCAAAAGGCGAGAAAATAATGGGTTTGGTACGCGAAGGGTCGTTATCTAGCTCAGTGGAAGCTGATCCAGATCTAACGTGGTCAGTACCTGAACACTGGACGCTTGAAGACGCAGCCACCGTTCCCCTAGCGTATCTTCATGCCTTCTATTGCTTGGCACTAAGGAGCAATGTTTCTAGTGGAAAAACAGTATTCATCACGGGAGGCGCTGGGGCTCTGGGACAAGCAGTCATCTCCATATGCCTGGCTATGAACTGTTCTATTTACACAACCGTAAAAGATATTTATAAGAAGAACTTTTTATTGCAGCTATTCCCTGAACTTCCAGAAAGTCACATCGGATTTTCGCACCTTGAAGCTTTATATAACATGCCTGTATTTAATTCTGAGAATAAACGTTGCGATATAATTATAAATTTAGACAGCGGTCATCTACGAGAGGCTTTGATGAAATGTGTCAGAGCCAGTGGCATATTTTTGGACATCAGCAAATCAGACATGAATAAAAACAAAGATTTCTACATGGGGTTTATCCGAGGCGATAGATCGTATAAGGCTGTTGATTTTTCTAATATTTTCAGGATTGAATTCGCTGAAGACAAGAAGGTATTTTTATGA
Protein
MDFSGYNSKGEKIMGLVREGSLSSSVEADPDLTWSVPEHWTLEDAATVPLAYLHAFYCLALRSNVSSGKTVFITGGAGALGQAVISICLAMNCSIYTTVKDIYKKNFLLQLFPELPESHIGFSHLEALYNMPVFNSENKRCDIIINLDSGHLREALMKCVRASGIFLDISKSDMNKNKDFYMGFIRGDRSYKAVDFSNIFRIEFAEDKKVFL
Summary
Uniprot
H9JGD2
A0A2A4IY92
H9JGF5
O01677
A0A2A4IZ90
A0A2H1V933
+ More
A0A3S2NJM7 A0A2H1WZR1 A0A194QVD5 A0A2A4JVR3 A0A2A4K4K9 A0A0L7LNK7 A0A2A4JTZ4 A0A2A4K3Y0 A0A2A4K3Y5 A0A194PKD7 A0A0N1I4Z6 A0A2H1VBX5 A0A2H1VPQ1 A0A212EWU6 A0A0L7KRD1 A0A2J7PFW9 A0A3S2M8J0 A0A0L7LPX1 A0A194QF04 A0A336M5W9 V5GRN6 A0A2J7PFW7 A0A1B2M4Y5 A0A1W4XJ56 A0A1Y1K5D2 K7IM26 A0A067RHL3 N6TYU8 U4UFT3 A0A2B4RJ56 A0A2J7PFF5 A0A344X1T4 A0A2D3E2Z1 A0A2R7X5T9 A0A232F3H2 A0A336T938 A0A0J7KN36 A0A2R7VR19 A0A224X4N0 A0A2J7Q8D1 A0A3M6U464 A0A0L7LTQ0 A0A2J7Q8C4 A0A2J7Q8C2 A0A1W7RAJ9 A0A2J7PFF1 A0A0J7MWI1 A0A2C9PHR5 A0A023GME4 A0A1W5YLL8 A0A1Y1MBP7 H3CK37 Q4SXH3 A0A1B6K868 A0A023GMF0 T1H8C9 A0A3R7M6P7 F8RHR0 A0A151I5V4 T1JE63 A0A1E1X9M5 D6W6D2 A0A310S575 D2A4M5 A0A147B8U8 A0A088AP68 A0A1B6MRN5 A0A0M9A0W5 A0A1B6GY38 A0A2A4K5B4 A0A026WXS0 A0A195DQH9 A0A3L8E0E3 A0A2I4CMN6 A0A158NAI9 A0A151X557 A0A1B6GNL2 A0A1S4F133 F4X257 A0A195CLZ5 E2AU37 A0A195EVF8 T1HFT7 A0A0J7L122 A0A182GVQ5 L7ML13 A0A194Q9W3 A0A232EXC2 E9JC01 A0A194QPH6 A0A0K0EY58
A0A3S2NJM7 A0A2H1WZR1 A0A194QVD5 A0A2A4JVR3 A0A2A4K4K9 A0A0L7LNK7 A0A2A4JTZ4 A0A2A4K3Y0 A0A2A4K3Y5 A0A194PKD7 A0A0N1I4Z6 A0A2H1VBX5 A0A2H1VPQ1 A0A212EWU6 A0A0L7KRD1 A0A2J7PFW9 A0A3S2M8J0 A0A0L7LPX1 A0A194QF04 A0A336M5W9 V5GRN6 A0A2J7PFW7 A0A1B2M4Y5 A0A1W4XJ56 A0A1Y1K5D2 K7IM26 A0A067RHL3 N6TYU8 U4UFT3 A0A2B4RJ56 A0A2J7PFF5 A0A344X1T4 A0A2D3E2Z1 A0A2R7X5T9 A0A232F3H2 A0A336T938 A0A0J7KN36 A0A2R7VR19 A0A224X4N0 A0A2J7Q8D1 A0A3M6U464 A0A0L7LTQ0 A0A2J7Q8C4 A0A2J7Q8C2 A0A1W7RAJ9 A0A2J7PFF1 A0A0J7MWI1 A0A2C9PHR5 A0A023GME4 A0A1W5YLL8 A0A1Y1MBP7 H3CK37 Q4SXH3 A0A1B6K868 A0A023GMF0 T1H8C9 A0A3R7M6P7 F8RHR0 A0A151I5V4 T1JE63 A0A1E1X9M5 D6W6D2 A0A310S575 D2A4M5 A0A147B8U8 A0A088AP68 A0A1B6MRN5 A0A0M9A0W5 A0A1B6GY38 A0A2A4K5B4 A0A026WXS0 A0A195DQH9 A0A3L8E0E3 A0A2I4CMN6 A0A158NAI9 A0A151X557 A0A1B6GNL2 A0A1S4F133 F4X257 A0A195CLZ5 E2AU37 A0A195EVF8 T1HFT7 A0A0J7L122 A0A182GVQ5 L7ML13 A0A194Q9W3 A0A232EXC2 E9JC01 A0A194QPH6 A0A0K0EY58
Pubmed
EMBL
BABH01033622
BABH01033623
BABH01033624
NWSH01005646
PCG64083.1
BABH01033637
+ More
U67866 AAB53257.1 NWSH01005017 PCG64490.1 ODYU01001294 SOQ37311.1 RSAL01000068 RVE49259.1 ODYU01012325 SOQ58579.1 KQ461073 KPJ09498.1 NWSH01000591 PCG75472.1 NWSH01000156 PCG78966.1 JTDY01000529 KOB76791.1 PCG75471.1 PCG78965.1 PCG78967.1 KQ459602 KPI93194.1 KQ461178 KPJ07769.1 ODYU01001484 SOQ37744.1 ODYU01003708 SOQ42815.1 AGBW02011888 OWR45966.1 JTDY01006902 KOB65606.1 NEVH01025646 PNF15231.1 RSAL01000013 RVE53445.1 JTDY01000426 KOB77236.1 KQ459053 KPJ04123.1 UFQT01000602 SSX25625.1 GALX01004174 JAB64292.1 PNF15229.1 KX467777 AOA60273.1 GEZM01096807 JAV54566.1 KK852680 KDR18643.1 APGK01049539 APGK01049540 KB741156 ENN73526.1 KB632169 ERL89446.1 LSMT01000411 PFX18424.1 NEVH01025661 PNF15062.1 MF817713 AXE71619.1 MF062033 ATU31751.1 KK857421 PTY27158.1 NNAY01001061 OXU25291.1 KM042200 AJW76499.1 LBMM01005284 KMQ91654.1 KK854036 PTY09977.1 GFTR01008959 JAW07467.1 NEVH01016965 PNF24835.1 RCHS01002288 RMX48359.1 JTDY01000102 KOB78858.1 PNF24833.1 PNF24834.1 GFAH01000232 JAV48157.1 PNF15063.1 LBMM01015399 KMQ84825.1 KY797274 ATL76713.1 GBBM01000412 JAC35006.1 KX147671 ARI45075.1 GEZM01035401 JAV83262.1 CAAE01012412 CAF94659.1 GECU01000102 JAT07605.1 GBBM01000409 JAC35009.1 ACPB03012542 QCYY01002654 ROT68596.1 HM595630 ADM88556.1 KQ976413 KYM90403.1 JH432116 GFAC01003246 JAT95942.1 KQ971319 EFA11365.1 KQ770827 OAD52584.1 KQ971344 EFA05203.2 GEIB01000832 JAR87184.1 GEBQ01001399 JAT38578.1 KQ435796 KOX73549.1 GECZ01002433 JAS67336.1 PCG78963.1 KK107077 QOIP01000010 EZA60541.1 RLU17791.1 KQ980612 KYN15123.1 QOIP01000002 RLU26096.1 ADTU01010333 ADTU01010334 KQ982519 KYQ55543.1 GECZ01005755 JAS64014.1 GL888565 EGI59473.1 KQ977642 KYN01094.1 GL442783 EFN63051.1 KQ981954 KYN32228.1 ACPB03012543 ACPB03012544 LBMM01001339 KMQ96487.1 JXUM01091581 JXUM01091582 GACK01000522 JAA64512.1 KQ459252 KPJ02209.1 NNAY01001760 OXU23005.1 GL771642 EFZ09693.1 KQ461185 KPJ07407.1
U67866 AAB53257.1 NWSH01005017 PCG64490.1 ODYU01001294 SOQ37311.1 RSAL01000068 RVE49259.1 ODYU01012325 SOQ58579.1 KQ461073 KPJ09498.1 NWSH01000591 PCG75472.1 NWSH01000156 PCG78966.1 JTDY01000529 KOB76791.1 PCG75471.1 PCG78965.1 PCG78967.1 KQ459602 KPI93194.1 KQ461178 KPJ07769.1 ODYU01001484 SOQ37744.1 ODYU01003708 SOQ42815.1 AGBW02011888 OWR45966.1 JTDY01006902 KOB65606.1 NEVH01025646 PNF15231.1 RSAL01000013 RVE53445.1 JTDY01000426 KOB77236.1 KQ459053 KPJ04123.1 UFQT01000602 SSX25625.1 GALX01004174 JAB64292.1 PNF15229.1 KX467777 AOA60273.1 GEZM01096807 JAV54566.1 KK852680 KDR18643.1 APGK01049539 APGK01049540 KB741156 ENN73526.1 KB632169 ERL89446.1 LSMT01000411 PFX18424.1 NEVH01025661 PNF15062.1 MF817713 AXE71619.1 MF062033 ATU31751.1 KK857421 PTY27158.1 NNAY01001061 OXU25291.1 KM042200 AJW76499.1 LBMM01005284 KMQ91654.1 KK854036 PTY09977.1 GFTR01008959 JAW07467.1 NEVH01016965 PNF24835.1 RCHS01002288 RMX48359.1 JTDY01000102 KOB78858.1 PNF24833.1 PNF24834.1 GFAH01000232 JAV48157.1 PNF15063.1 LBMM01015399 KMQ84825.1 KY797274 ATL76713.1 GBBM01000412 JAC35006.1 KX147671 ARI45075.1 GEZM01035401 JAV83262.1 CAAE01012412 CAF94659.1 GECU01000102 JAT07605.1 GBBM01000409 JAC35009.1 ACPB03012542 QCYY01002654 ROT68596.1 HM595630 ADM88556.1 KQ976413 KYM90403.1 JH432116 GFAC01003246 JAT95942.1 KQ971319 EFA11365.1 KQ770827 OAD52584.1 KQ971344 EFA05203.2 GEIB01000832 JAR87184.1 GEBQ01001399 JAT38578.1 KQ435796 KOX73549.1 GECZ01002433 JAS67336.1 PCG78963.1 KK107077 QOIP01000010 EZA60541.1 RLU17791.1 KQ980612 KYN15123.1 QOIP01000002 RLU26096.1 ADTU01010333 ADTU01010334 KQ982519 KYQ55543.1 GECZ01005755 JAS64014.1 GL888565 EGI59473.1 KQ977642 KYN01094.1 GL442783 EFN63051.1 KQ981954 KYN32228.1 ACPB03012543 ACPB03012544 LBMM01001339 KMQ96487.1 JXUM01091581 JXUM01091582 GACK01000522 JAA64512.1 KQ459252 KPJ02209.1 NNAY01001760 OXU23005.1 GL771642 EFZ09693.1 KQ461185 KPJ07407.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000037510
UP000053268
+ More
UP000007151 UP000235965 UP000192223 UP000002358 UP000027135 UP000019118 UP000030742 UP000225706 UP000215335 UP000036403 UP000275408 UP000007303 UP000015103 UP000283509 UP000078540 UP000007266 UP000005203 UP000053105 UP000053097 UP000279307 UP000078492 UP000192220 UP000005205 UP000075809 UP000007755 UP000078542 UP000000311 UP000078541 UP000069940 UP000035680
UP000007151 UP000235965 UP000192223 UP000002358 UP000027135 UP000019118 UP000030742 UP000225706 UP000215335 UP000036403 UP000275408 UP000007303 UP000015103 UP000283509 UP000078540 UP000007266 UP000005203 UP000053105 UP000053097 UP000279307 UP000078492 UP000192220 UP000005205 UP000075809 UP000007755 UP000078542 UP000000311 UP000078541 UP000069940 UP000035680
Pfam
Interpro
IPR020841
PKS_Beta-ketoAc_synthase_dom
+ More
IPR020846 MFS_dom
IPR001227 Ac_transferase_dom_sf
IPR036259 MFS_trans_sf
IPR020843 PKS_ER
IPR014030 Ketoacyl_synth_N
IPR016039 Thiolase-like
IPR020801 PKS_acyl_transferase
IPR011701 MFS
IPR042104 PKS_dehydratase_sf
IPR011032 GroES-like_sf
IPR036291 NAD(P)-bd_dom_sf
IPR014043 Acyl_transferase
IPR016036 Malonyl_transacylase_ACP-bd
IPR016035 Acyl_Trfase/lysoPLipase
IPR014031 Ketoacyl_synth_C
IPR013968 PKS_KR
IPR020807 PKS_dehydratase
IPR029058 AB_hydrolase
IPR023102 Fatty_acid_synthase_dom_2
IPR032821 KAsynt_C_assoc
IPR001031 Thioesterase
IPR018201 Ketoacyl_synth_AS
IPR013149 ADH_C
IPR020806 PKS_PP-bd
IPR009081 PP-bd_ACP
IPR036736 ACP-like_sf
IPR006162 Ppantetheine_attach_site
IPR029063 SAM-dependent_MTases
IPR020802 PKS_thioesterase
IPR020846 MFS_dom
IPR001227 Ac_transferase_dom_sf
IPR036259 MFS_trans_sf
IPR020843 PKS_ER
IPR014030 Ketoacyl_synth_N
IPR016039 Thiolase-like
IPR020801 PKS_acyl_transferase
IPR011701 MFS
IPR042104 PKS_dehydratase_sf
IPR011032 GroES-like_sf
IPR036291 NAD(P)-bd_dom_sf
IPR014043 Acyl_transferase
IPR016036 Malonyl_transacylase_ACP-bd
IPR016035 Acyl_Trfase/lysoPLipase
IPR014031 Ketoacyl_synth_C
IPR013968 PKS_KR
IPR020807 PKS_dehydratase
IPR029058 AB_hydrolase
IPR023102 Fatty_acid_synthase_dom_2
IPR032821 KAsynt_C_assoc
IPR001031 Thioesterase
IPR018201 Ketoacyl_synth_AS
IPR013149 ADH_C
IPR020806 PKS_PP-bd
IPR009081 PP-bd_ACP
IPR036736 ACP-like_sf
IPR006162 Ppantetheine_attach_site
IPR029063 SAM-dependent_MTases
IPR020802 PKS_thioesterase
SUPFAM
CDD
ProteinModelPortal
H9JGD2
A0A2A4IY92
H9JGF5
O01677
A0A2A4IZ90
A0A2H1V933
+ More
A0A3S2NJM7 A0A2H1WZR1 A0A194QVD5 A0A2A4JVR3 A0A2A4K4K9 A0A0L7LNK7 A0A2A4JTZ4 A0A2A4K3Y0 A0A2A4K3Y5 A0A194PKD7 A0A0N1I4Z6 A0A2H1VBX5 A0A2H1VPQ1 A0A212EWU6 A0A0L7KRD1 A0A2J7PFW9 A0A3S2M8J0 A0A0L7LPX1 A0A194QF04 A0A336M5W9 V5GRN6 A0A2J7PFW7 A0A1B2M4Y5 A0A1W4XJ56 A0A1Y1K5D2 K7IM26 A0A067RHL3 N6TYU8 U4UFT3 A0A2B4RJ56 A0A2J7PFF5 A0A344X1T4 A0A2D3E2Z1 A0A2R7X5T9 A0A232F3H2 A0A336T938 A0A0J7KN36 A0A2R7VR19 A0A224X4N0 A0A2J7Q8D1 A0A3M6U464 A0A0L7LTQ0 A0A2J7Q8C4 A0A2J7Q8C2 A0A1W7RAJ9 A0A2J7PFF1 A0A0J7MWI1 A0A2C9PHR5 A0A023GME4 A0A1W5YLL8 A0A1Y1MBP7 H3CK37 Q4SXH3 A0A1B6K868 A0A023GMF0 T1H8C9 A0A3R7M6P7 F8RHR0 A0A151I5V4 T1JE63 A0A1E1X9M5 D6W6D2 A0A310S575 D2A4M5 A0A147B8U8 A0A088AP68 A0A1B6MRN5 A0A0M9A0W5 A0A1B6GY38 A0A2A4K5B4 A0A026WXS0 A0A195DQH9 A0A3L8E0E3 A0A2I4CMN6 A0A158NAI9 A0A151X557 A0A1B6GNL2 A0A1S4F133 F4X257 A0A195CLZ5 E2AU37 A0A195EVF8 T1HFT7 A0A0J7L122 A0A182GVQ5 L7ML13 A0A194Q9W3 A0A232EXC2 E9JC01 A0A194QPH6 A0A0K0EY58
A0A3S2NJM7 A0A2H1WZR1 A0A194QVD5 A0A2A4JVR3 A0A2A4K4K9 A0A0L7LNK7 A0A2A4JTZ4 A0A2A4K3Y0 A0A2A4K3Y5 A0A194PKD7 A0A0N1I4Z6 A0A2H1VBX5 A0A2H1VPQ1 A0A212EWU6 A0A0L7KRD1 A0A2J7PFW9 A0A3S2M8J0 A0A0L7LPX1 A0A194QF04 A0A336M5W9 V5GRN6 A0A2J7PFW7 A0A1B2M4Y5 A0A1W4XJ56 A0A1Y1K5D2 K7IM26 A0A067RHL3 N6TYU8 U4UFT3 A0A2B4RJ56 A0A2J7PFF5 A0A344X1T4 A0A2D3E2Z1 A0A2R7X5T9 A0A232F3H2 A0A336T938 A0A0J7KN36 A0A2R7VR19 A0A224X4N0 A0A2J7Q8D1 A0A3M6U464 A0A0L7LTQ0 A0A2J7Q8C4 A0A2J7Q8C2 A0A1W7RAJ9 A0A2J7PFF1 A0A0J7MWI1 A0A2C9PHR5 A0A023GME4 A0A1W5YLL8 A0A1Y1MBP7 H3CK37 Q4SXH3 A0A1B6K868 A0A023GMF0 T1H8C9 A0A3R7M6P7 F8RHR0 A0A151I5V4 T1JE63 A0A1E1X9M5 D6W6D2 A0A310S575 D2A4M5 A0A147B8U8 A0A088AP68 A0A1B6MRN5 A0A0M9A0W5 A0A1B6GY38 A0A2A4K5B4 A0A026WXS0 A0A195DQH9 A0A3L8E0E3 A0A2I4CMN6 A0A158NAI9 A0A151X557 A0A1B6GNL2 A0A1S4F133 F4X257 A0A195CLZ5 E2AU37 A0A195EVF8 T1HFT7 A0A0J7L122 A0A182GVQ5 L7ML13 A0A194Q9W3 A0A232EXC2 E9JC01 A0A194QPH6 A0A0K0EY58
PDB
4W9N
E-value=2.92798e-33,
Score=351
Ontologies
GO
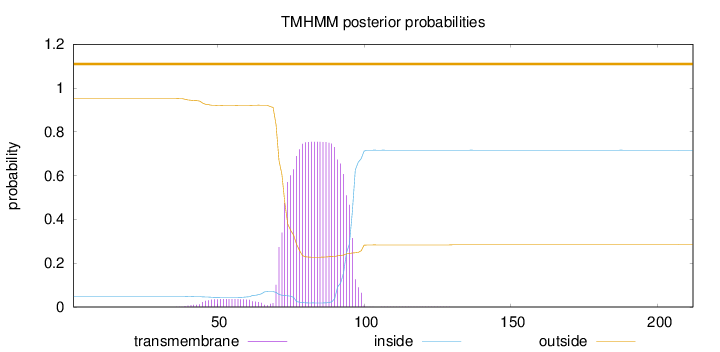
Topology
Length:
212
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
17.68483
Exp number, first 60 AAs:
0.59121
Total prob of N-in:
0.04799
outside
1 - 212
Population Genetic Test Statistics
Pi
19.432033
Theta
24.04162
Tajima's D
-0.544998
CLR
3.193215
CSRT
0.230238488075596
Interpretation
Uncertain
