Pre Gene Modal
BGIBMGA008602
Annotation
p260_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.716 Nuclear Reliability : 1.233
Sequence
CDS
ATGGTAATACAATATGATTGCTCTTTCAGATGTGGTGGAGTTGAATTAGTTGGACTAACGTTCAACGATAAACCAGCAGTTGAAAAAGAACCTGATGTTATGCAATCTAGAGTCTTTGTTCCATACTTTGTAACTGGAGTAGTTGATCTCAAGACTGCGTTACAAGTCTCTTTTGGAACCATAAGTGACAATGTAAATGGCGAGGAAGTCAATGTGATCGAATGCTTGAGATCAGAAGACCACGAAATCTCAAAGCTCATCAAGGAAGTGACAAGAACACATGCAAGCGCTAATTTCGACGTTGTTTCTGTTGAATCATTAGATTTGAAAGCTGTTAGCGGAGCAGATGTCCTGATTACTGTTAATTTGTTAGGAAACGAAATGGAAATGAAGAAAATACATGATAGTCTACGCCAGAATAGTTTCATCCTATCCATGGAAGAAGACATTAGTAATATCAATCCGGATCACTCTCTTTTCACAACTGTATCGGCCATGTCCGTTCAGAACCATATATTGGTTTTGTTGCGCAAGACCTCGGAGGATATTGATGTTACGTTTATGCCTATCAATCACGACATCAACTTCACTTGGGTATCACGGTTGAGGAACGAAATCGAAAAATCAAGAAAGGTCGTTTTGATTTCAGAGAGACAACCCTATTGCGGACTTTACGGTTTAGTTAAGAAACTAAGTAAGCAACCGAGAAATAATGTAAGCTTAGTACTGGTTGACGACTTCTACCATGTTCCATCATTTGATGCTGAGCATCCACTGTTCAAAGAACAACTACGGAAGAACTTGACTTTCAACGTTTTGAAAAAGGGTGTATGGGGCAGCTTCTACTATTTGTCCAATGATTCATACTCGAACTTCCAAAACGTTTACTTGACCAATGCTATTACCGGTGACTTGAACAGCCTAACTTGGATTGAGGCATCATCTACTCCGGCCAATAATAACTTAGTTCAGGTCTGCTACAGTAGCCCGTCGTACAGAGATGCACAAGATGCCGCTGGAACCACATCTAAGTCCAACAGATCATTTGGAATGGATTACAGTGGTATTAACAGTCTGGGTGAAAGGGTAATGGGTGTAGTACAGGGAGGAGCTCTGTCGACTTTGGTGGAAGCTGACCCGGATCTAACGTGGCCGGTACCTGAACACTGGACTCTCGAAGACGCTGCCACTGTGCCTTTACCATATCTACAAGCTTATTATTGTTTAGCAATTAAATCTGAATTATTGAGAAAAAGAAAAGTTTTTATCAATGGAGGAGCCGGAGCATTAGGTCAAGCCGTCATTTCAATCTGTCTCGCTTTGGATTGTGAGGTTTTCACGACAGTAAGCGACACGAAAAAGAAAGAATTCCTAATGAAGTTGTTCCCGAGTTTGCCGGAATCGAATATTGCAAGCTCCCATAATATTCATTTCAAGGATAGGGTCATGCAGAACACTAAAAATAAAGGATGTGACGTCGTCATCAATTACATGAACGGAAATCTACGTGAGGTTATCGTTATAGTGCCATCAGTATACAATTAA
Protein
MVIQYDCSFRCGGVELVGLTFNDKPAVEKEPDVMQSRVFVPYFVTGVVDLKTALQVSFGTISDNVNGEEVNVIECLRSEDHEISKLIKEVTRTHASANFDVVSVESLDLKAVSGADVLITVNLLGNEMEMKKIHDSLRQNSFILSMEEDISNINPDHSLFTTVSAMSVQNHILVLLRKTSEDIDVTFMPINHDINFTWVSRLRNEIEKSRKVVLISERQPYCGLYGLVKKLSKQPRNNVSLVLVDDFYHVPSFDAEHPLFKEQLRKNLTFNVLKKGVWGSFYYLSNDSYSNFQNVYLTNAITGDLNSLTWIEASSTPANNNLVQVCYSSPSYRDAQDAAGTTSKSNRSFGMDYSGINSLGERVMGVVQGGALSTLVEADPDLTWPVPEHWTLEDAATVPLPYLQAYYCLAIKSELLRKRKVFINGGAGALGQAVISICLALDCEVFTTVSDTKKKEFLMKLFPSLPESNIASSHNIHFKDRVMQNTKNKGCDVVINYMNGNLREVIVIVPSVYN
Summary
Uniprot
H9JGF5
O01677
A0A3S2NJM7
A0A2A4JVR3
A0A2A4IZ90
A0A2H1V933
+ More
A0A2H1WZR1 A0A2A4IY92 A0A2A4JTZ4 H9JGD4 A0A0N1I4Z6 A0A3S2M8J0 A0A2A4K3Y5 A0A2A4K4K9 A0A194PKD7 A0A212EWU6 A0A336L600 A0A2H1VBX5 A0A067RHL3 A0A2P8XAG4 A0A0L7L9U1 A0A2P8XIL1 A0A1Q3FS82 A0A2H1VFB6 A0A2H1VPQ1 B0WMC9 A0A2H1WSB0 Q16ZI9 A0A1S4FIT0 A0A026WJ58 A0A182G1R8 A0A2J7PFW7 A0A0L7LTQ0 A0A3L8DZ17 A0A084VRX8 A0A182GNP7 A0A3L8DXI6 H9JU35 A0A336N0W1 A0A1W4VE27 A0A0J7NXT7 W5J299 A0A0L7L2S0 A0A087ZKN3 A0A026WFH9 A0A182FQK1 A0A2H1WKG4 O01678 A0A336MVQ6 A0A212FMU5 A0A336MX78 A0A1S4F133 A0A1Q3FRQ3 A0A026WIF7 A0A336MXS7 A0A3L8DDP5 A0A2H1WWR5 A0A336M5W9 B4NIW4 A0A224X4N0 A0A1J1IKA4 Q17IW6 A0A151X6P9 A0A2H1W5A9 A0A182M3X3 A0A1B6CY84 A0A2H8U0Z2 A0A1B6DTB3 A0A195F360 A0A1B6D462 N6T056 U4TX81 A0A0N1IP09 D2A4M5 B0W8R9 A0A0G3YL52 N6TW05 A0A182PC27 A0A182R1J1 A0A182GVQ5 J9K5E2 A0A182JQR8 A0A0M4EN44 A0A2W1BHS7 A0A195CP13 A0A194Q6Q7 A0A182VQY2 A0A2S2RA89 A0A151IJ25 A0A232EXB1 A0A182MEL3 E2ADN8 A0A2A4J2W6 G7H848 A0A1Q3FSZ4 D2NUK3 A0A0J7KN36 A0A2A4J169 T1E9A6
A0A2H1WZR1 A0A2A4IY92 A0A2A4JTZ4 H9JGD4 A0A0N1I4Z6 A0A3S2M8J0 A0A2A4K3Y5 A0A2A4K4K9 A0A194PKD7 A0A212EWU6 A0A336L600 A0A2H1VBX5 A0A067RHL3 A0A2P8XAG4 A0A0L7L9U1 A0A2P8XIL1 A0A1Q3FS82 A0A2H1VFB6 A0A2H1VPQ1 B0WMC9 A0A2H1WSB0 Q16ZI9 A0A1S4FIT0 A0A026WJ58 A0A182G1R8 A0A2J7PFW7 A0A0L7LTQ0 A0A3L8DZ17 A0A084VRX8 A0A182GNP7 A0A3L8DXI6 H9JU35 A0A336N0W1 A0A1W4VE27 A0A0J7NXT7 W5J299 A0A0L7L2S0 A0A087ZKN3 A0A026WFH9 A0A182FQK1 A0A2H1WKG4 O01678 A0A336MVQ6 A0A212FMU5 A0A336MX78 A0A1S4F133 A0A1Q3FRQ3 A0A026WIF7 A0A336MXS7 A0A3L8DDP5 A0A2H1WWR5 A0A336M5W9 B4NIW4 A0A224X4N0 A0A1J1IKA4 Q17IW6 A0A151X6P9 A0A2H1W5A9 A0A182M3X3 A0A1B6CY84 A0A2H8U0Z2 A0A1B6DTB3 A0A195F360 A0A1B6D462 N6T056 U4TX81 A0A0N1IP09 D2A4M5 B0W8R9 A0A0G3YL52 N6TW05 A0A182PC27 A0A182R1J1 A0A182GVQ5 J9K5E2 A0A182JQR8 A0A0M4EN44 A0A2W1BHS7 A0A195CP13 A0A194Q6Q7 A0A182VQY2 A0A2S2RA89 A0A151IJ25 A0A232EXB1 A0A182MEL3 E2ADN8 A0A2A4J2W6 G7H848 A0A1Q3FSZ4 D2NUK3 A0A0J7KN36 A0A2A4J169 T1E9A6
Pubmed
EMBL
BABH01033637
U67866
AAB53257.1
RSAL01000068
RVE49259.1
NWSH01000591
+ More
PCG75472.1 NWSH01005017 PCG64490.1 ODYU01001294 SOQ37311.1 ODYU01012325 SOQ58579.1 NWSH01005646 PCG64083.1 PCG75471.1 BABH01033613 BABH01033614 BABH01033615 BABH01033616 BABH01033617 BABH01033618 KQ461178 KPJ07769.1 RSAL01000013 RVE53445.1 NWSH01000156 PCG78967.1 PCG78966.1 KQ459602 KPI93194.1 AGBW02011888 OWR45966.1 UFQS01001117 UFQT01001117 SSX09070.1 SSX28981.1 ODYU01001484 SOQ37744.1 KK852680 KDR18643.1 PYGN01016841 PSN28987.1 JTDY01002046 KOB72242.1 PYGN01002008 PSN31835.1 GFDL01004709 JAV30336.1 ODYU01002245 SOQ39471.1 ODYU01003708 SOQ42815.1 DS231997 EDS30986.1 ODYU01010623 SOQ55856.1 CH477489 EAT40085.1 KK107182 EZA56033.1 JXUM01038912 KQ561184 KXJ79436.1 NEVH01025646 PNF15229.1 JTDY01000102 KOB78858.1 QOIP01000003 RLU25189.1 ATLV01015783 ATLV01015784 KE525036 KFB40722.1 JXUM01076840 KQ562972 KXJ74732.1 RLU25184.1 BABH01003987 BABH01003988 UFQT01002952 SSX34337.1 LBMM01000911 KMQ97230.1 ADMH02002134 ETN58382.1 JTDY01003335 KOB69710.1 KK107273 EZA53779.1 ODYU01009273 SOQ53573.1 U67867 AAB53258.1 SSX34336.1 AGBW02007651 OWR55047.1 SSX34335.1 GFDL01004798 JAV30247.1 KK107235 EZA54889.1 UFQT01003008 SSX34421.1 QOIP01000009 RLU18610.1 ODYU01011246 SOQ56864.1 UFQT01000602 SSX25625.1 CH964272 EDW84866.1 GFTR01008959 JAW07467.1 CVRI01000051 CRK99510.1 CH477236 EAT46627.1 KQ982480 KYQ55999.1 ODYU01006431 SOQ48269.1 AXCM01001692 GEDC01031101 GEDC01028079 GEDC01018856 GEDC01013285 JAS06197.1 JAS09219.1 JAS18442.1 JAS24013.1 GFXV01008055 MBW19860.1 GEDC01031722 GEDC01008387 JAS05576.1 JAS28911.1 KQ981856 KYN34617.1 GEDC01016862 GEDC01012243 JAS20436.1 JAS25055.1 APGK01057373 KB741280 ENN70888.1 KB631604 ERL84608.1 KQ460651 KPJ13003.1 KQ971344 EFA05203.2 DS231860 EDS39298.1 KM507100 AKM28424.1 APGK01052936 APGK01052937 KB741216 ENN72556.1 JXUM01091581 JXUM01091582 ABLF02040018 CP012526 ALC46026.1 KZ150116 PZC73265.1 KQ977574 KYN01849.1 KQ459582 KPI99075.1 GGMS01017661 MBY86864.1 KQ977412 KYN02883.1 NNAY01001760 OXU23004.1 AXCM01003979 GL438820 EFN68400.1 NWSH01003873 PCG65733.1 BT132772 AET14856.1 GFDL01004326 JAV30719.1 BT120146 ADB57067.1 LBMM01005284 KMQ91654.1 PCG65731.1 GAMD01002013 JAA99577.1
PCG75472.1 NWSH01005017 PCG64490.1 ODYU01001294 SOQ37311.1 ODYU01012325 SOQ58579.1 NWSH01005646 PCG64083.1 PCG75471.1 BABH01033613 BABH01033614 BABH01033615 BABH01033616 BABH01033617 BABH01033618 KQ461178 KPJ07769.1 RSAL01000013 RVE53445.1 NWSH01000156 PCG78967.1 PCG78966.1 KQ459602 KPI93194.1 AGBW02011888 OWR45966.1 UFQS01001117 UFQT01001117 SSX09070.1 SSX28981.1 ODYU01001484 SOQ37744.1 KK852680 KDR18643.1 PYGN01016841 PSN28987.1 JTDY01002046 KOB72242.1 PYGN01002008 PSN31835.1 GFDL01004709 JAV30336.1 ODYU01002245 SOQ39471.1 ODYU01003708 SOQ42815.1 DS231997 EDS30986.1 ODYU01010623 SOQ55856.1 CH477489 EAT40085.1 KK107182 EZA56033.1 JXUM01038912 KQ561184 KXJ79436.1 NEVH01025646 PNF15229.1 JTDY01000102 KOB78858.1 QOIP01000003 RLU25189.1 ATLV01015783 ATLV01015784 KE525036 KFB40722.1 JXUM01076840 KQ562972 KXJ74732.1 RLU25184.1 BABH01003987 BABH01003988 UFQT01002952 SSX34337.1 LBMM01000911 KMQ97230.1 ADMH02002134 ETN58382.1 JTDY01003335 KOB69710.1 KK107273 EZA53779.1 ODYU01009273 SOQ53573.1 U67867 AAB53258.1 SSX34336.1 AGBW02007651 OWR55047.1 SSX34335.1 GFDL01004798 JAV30247.1 KK107235 EZA54889.1 UFQT01003008 SSX34421.1 QOIP01000009 RLU18610.1 ODYU01011246 SOQ56864.1 UFQT01000602 SSX25625.1 CH964272 EDW84866.1 GFTR01008959 JAW07467.1 CVRI01000051 CRK99510.1 CH477236 EAT46627.1 KQ982480 KYQ55999.1 ODYU01006431 SOQ48269.1 AXCM01001692 GEDC01031101 GEDC01028079 GEDC01018856 GEDC01013285 JAS06197.1 JAS09219.1 JAS18442.1 JAS24013.1 GFXV01008055 MBW19860.1 GEDC01031722 GEDC01008387 JAS05576.1 JAS28911.1 KQ981856 KYN34617.1 GEDC01016862 GEDC01012243 JAS20436.1 JAS25055.1 APGK01057373 KB741280 ENN70888.1 KB631604 ERL84608.1 KQ460651 KPJ13003.1 KQ971344 EFA05203.2 DS231860 EDS39298.1 KM507100 AKM28424.1 APGK01052936 APGK01052937 KB741216 ENN72556.1 JXUM01091581 JXUM01091582 ABLF02040018 CP012526 ALC46026.1 KZ150116 PZC73265.1 KQ977574 KYN01849.1 KQ459582 KPI99075.1 GGMS01017661 MBY86864.1 KQ977412 KYN02883.1 NNAY01001760 OXU23004.1 AXCM01003979 GL438820 EFN68400.1 NWSH01003873 PCG65733.1 BT132772 AET14856.1 GFDL01004326 JAV30719.1 BT120146 ADB57067.1 LBMM01005284 KMQ91654.1 PCG65731.1 GAMD01002013 JAA99577.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053240
UP000053268
UP000007151
+ More
UP000027135 UP000245037 UP000037510 UP000002320 UP000008820 UP000053097 UP000069940 UP000249989 UP000235965 UP000279307 UP000030765 UP000192221 UP000036403 UP000000673 UP000069272 UP000007798 UP000183832 UP000075809 UP000075883 UP000078541 UP000019118 UP000030742 UP000007266 UP000075885 UP000075900 UP000007819 UP000075881 UP000092553 UP000078542 UP000075920 UP000215335 UP000000311
UP000027135 UP000245037 UP000037510 UP000002320 UP000008820 UP000053097 UP000069940 UP000249989 UP000235965 UP000279307 UP000030765 UP000192221 UP000036403 UP000000673 UP000069272 UP000007798 UP000183832 UP000075809 UP000075883 UP000078541 UP000019118 UP000030742 UP000007266 UP000075885 UP000075900 UP000007819 UP000075881 UP000092553 UP000078542 UP000075920 UP000215335 UP000000311
Pfam
Interpro
IPR016039
Thiolase-like
+ More
IPR020843 PKS_ER
IPR014030 Ketoacyl_synth_N
IPR020801 PKS_acyl_transferase
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR020807 PKS_dehydratase
IPR001227 Ac_transferase_dom_sf
IPR029058 AB_hydrolase
IPR016036 Malonyl_transacylase_ACP-bd
IPR014031 Ketoacyl_synth_C
IPR023102 Fatty_acid_synthase_dom_2
IPR016035 Acyl_Trfase/lysoPLipase
IPR032821 KAsynt_C_assoc
IPR042104 PKS_dehydratase_sf
IPR011032 GroES-like_sf
IPR001031 Thioesterase
IPR014043 Acyl_transferase
IPR036291 NAD(P)-bd_dom_sf
IPR018201 Ketoacyl_synth_AS
IPR013149 ADH_C
IPR013968 PKS_KR
IPR020806 PKS_PP-bd
IPR009081 PP-bd_ACP
IPR036736 ACP-like_sf
IPR006162 Ppantetheine_attach_site
IPR020843 PKS_ER
IPR014030 Ketoacyl_synth_N
IPR020801 PKS_acyl_transferase
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR020807 PKS_dehydratase
IPR001227 Ac_transferase_dom_sf
IPR029058 AB_hydrolase
IPR016036 Malonyl_transacylase_ACP-bd
IPR014031 Ketoacyl_synth_C
IPR023102 Fatty_acid_synthase_dom_2
IPR016035 Acyl_Trfase/lysoPLipase
IPR032821 KAsynt_C_assoc
IPR042104 PKS_dehydratase_sf
IPR011032 GroES-like_sf
IPR001031 Thioesterase
IPR014043 Acyl_transferase
IPR036291 NAD(P)-bd_dom_sf
IPR018201 Ketoacyl_synth_AS
IPR013149 ADH_C
IPR013968 PKS_KR
IPR020806 PKS_PP-bd
IPR009081 PP-bd_ACP
IPR036736 ACP-like_sf
IPR006162 Ppantetheine_attach_site
SUPFAM
ProteinModelPortal
H9JGF5
O01677
A0A3S2NJM7
A0A2A4JVR3
A0A2A4IZ90
A0A2H1V933
+ More
A0A2H1WZR1 A0A2A4IY92 A0A2A4JTZ4 H9JGD4 A0A0N1I4Z6 A0A3S2M8J0 A0A2A4K3Y5 A0A2A4K4K9 A0A194PKD7 A0A212EWU6 A0A336L600 A0A2H1VBX5 A0A067RHL3 A0A2P8XAG4 A0A0L7L9U1 A0A2P8XIL1 A0A1Q3FS82 A0A2H1VFB6 A0A2H1VPQ1 B0WMC9 A0A2H1WSB0 Q16ZI9 A0A1S4FIT0 A0A026WJ58 A0A182G1R8 A0A2J7PFW7 A0A0L7LTQ0 A0A3L8DZ17 A0A084VRX8 A0A182GNP7 A0A3L8DXI6 H9JU35 A0A336N0W1 A0A1W4VE27 A0A0J7NXT7 W5J299 A0A0L7L2S0 A0A087ZKN3 A0A026WFH9 A0A182FQK1 A0A2H1WKG4 O01678 A0A336MVQ6 A0A212FMU5 A0A336MX78 A0A1S4F133 A0A1Q3FRQ3 A0A026WIF7 A0A336MXS7 A0A3L8DDP5 A0A2H1WWR5 A0A336M5W9 B4NIW4 A0A224X4N0 A0A1J1IKA4 Q17IW6 A0A151X6P9 A0A2H1W5A9 A0A182M3X3 A0A1B6CY84 A0A2H8U0Z2 A0A1B6DTB3 A0A195F360 A0A1B6D462 N6T056 U4TX81 A0A0N1IP09 D2A4M5 B0W8R9 A0A0G3YL52 N6TW05 A0A182PC27 A0A182R1J1 A0A182GVQ5 J9K5E2 A0A182JQR8 A0A0M4EN44 A0A2W1BHS7 A0A195CP13 A0A194Q6Q7 A0A182VQY2 A0A2S2RA89 A0A151IJ25 A0A232EXB1 A0A182MEL3 E2ADN8 A0A2A4J2W6 G7H848 A0A1Q3FSZ4 D2NUK3 A0A0J7KN36 A0A2A4J169 T1E9A6
A0A2H1WZR1 A0A2A4IY92 A0A2A4JTZ4 H9JGD4 A0A0N1I4Z6 A0A3S2M8J0 A0A2A4K3Y5 A0A2A4K4K9 A0A194PKD7 A0A212EWU6 A0A336L600 A0A2H1VBX5 A0A067RHL3 A0A2P8XAG4 A0A0L7L9U1 A0A2P8XIL1 A0A1Q3FS82 A0A2H1VFB6 A0A2H1VPQ1 B0WMC9 A0A2H1WSB0 Q16ZI9 A0A1S4FIT0 A0A026WJ58 A0A182G1R8 A0A2J7PFW7 A0A0L7LTQ0 A0A3L8DZ17 A0A084VRX8 A0A182GNP7 A0A3L8DXI6 H9JU35 A0A336N0W1 A0A1W4VE27 A0A0J7NXT7 W5J299 A0A0L7L2S0 A0A087ZKN3 A0A026WFH9 A0A182FQK1 A0A2H1WKG4 O01678 A0A336MVQ6 A0A212FMU5 A0A336MX78 A0A1S4F133 A0A1Q3FRQ3 A0A026WIF7 A0A336MXS7 A0A3L8DDP5 A0A2H1WWR5 A0A336M5W9 B4NIW4 A0A224X4N0 A0A1J1IKA4 Q17IW6 A0A151X6P9 A0A2H1W5A9 A0A182M3X3 A0A1B6CY84 A0A2H8U0Z2 A0A1B6DTB3 A0A195F360 A0A1B6D462 N6T056 U4TX81 A0A0N1IP09 D2A4M5 B0W8R9 A0A0G3YL52 N6TW05 A0A182PC27 A0A182R1J1 A0A182GVQ5 J9K5E2 A0A182JQR8 A0A0M4EN44 A0A2W1BHS7 A0A195CP13 A0A194Q6Q7 A0A182VQY2 A0A2S2RA89 A0A151IJ25 A0A232EXB1 A0A182MEL3 E2ADN8 A0A2A4J2W6 G7H848 A0A1Q3FSZ4 D2NUK3 A0A0J7KN36 A0A2A4J169 T1E9A6
PDB
6FN6
E-value=1.22102e-47,
Score=480
Ontologies
GO
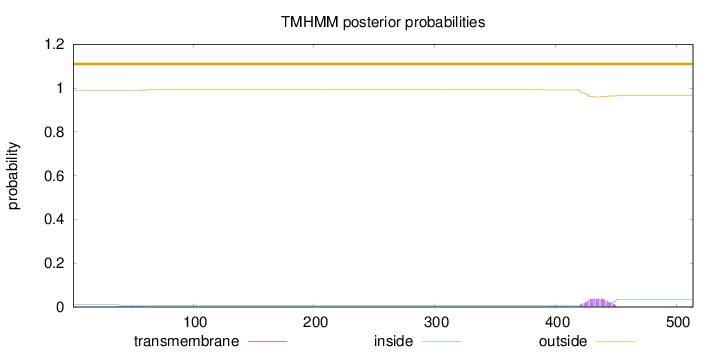
Topology
Length:
514
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.88009
Exp number, first 60 AAs:
0.04777
Total prob of N-in:
0.01061
outside
1 - 514
Population Genetic Test Statistics
Pi
14.351942
Theta
22.766131
Tajima's D
-1.107575
CLR
10.967167
CSRT
0.118394080295985
Interpretation
Uncertain
