Gene
KWMTBOMO11140
Annotation
p260_[Bombyx_mori]
Full name
Fatty acid synthase
Location in the cell
Nuclear Reliability : 1.146 PlasmaMembrane Reliability : 1.46
Sequence
CDS
ATGAGCTTTTTGTTTACATCCGGGAGCTATGTATCAACACAATTTTCTTCGATCTTCAAAACTGAAGGACTAAACGAGAAAAAGTTGCTGCACAAATTGATTAGCGAAGGTATAGCAAGTGGAGTGGTGAGGCCCCTCAGTCGGATGGTGTATTCTCCAGCGAATGTATCCAGAGCGTTCCGTCTCCTGGCCTCCAGTAAGCACCGAGGCAAGGTTTTGATTAAAATGAGGGACTCCAACACGAATCACGACTTGAAGGTTCATCCAAGATTGACCTGTACATCGAGGGGAAGCTATATCGTTGTCTGTGATGAAAGCGCTCTAAGCTTGGAACTGATTGATCGCTTAGTGAAACGTGGTGCCAGAAATCTGTTTATCCACATCAAGCCTAATTCTATCGCCGGATATTGTTTCACTAAAATTTTCAAGTGGAAAAAACTGAACGTGACAATCAAGATCTCGTCAGAAAACTTACAAACAGAGAACGGATGCACAAACATGATCAGAGAAGCGACTAAAACTGGACCAGTTTGTGGTATCTTCATAGTACAAAACTACAGTTCTGATGCCCAAGAAGATCAGTTCAGAGCTAATGAACATTTGTAA
Protein
MSFLFTSGSYVSTQFSSIFKTEGLNEKKLLHKLISEGIASGVVRPLSRMVYSPANVSRAFRLLASSKHRGKVLIKMRDSNTNHDLKVHPRLTCTSRGSYIVVCDESALSLELIDRLVKRGARNLFIHIKPNSIAGYCFTKIFKWKKLNVTIKISSENLQTENGCTNMIREATKTGPVCGIFIVQNYSSDAQEDQFRANEHL
Summary
Catalytic Activity
acetyl-CoA + 2n H(+) + n malonyl-CoA + 2n NADPH = a long-chain fatty acid + n CO2 + (n+1) CoA + 2n NADP(+)
acetyl-CoA + holo-[ACP] = acetyl-[ACP] + CoA
holo-[ACP] + malonyl-CoA = CoA + malonyl-[ACP]
a fatty acyl-[ACP] + H(+) + malonyl-[ACP] = a 3-oxoacyl-[ACP] + CO2 + holo-[ACP]
a (3R)-hydroxyacyl-[ACP] + NADP(+) = a 3-oxoacyl-[ACP] + H(+) + NADPH
a (3R)-hydroxyacyl-[ACP] = a (2E)-enoyl-[ACP] + H2O
a 2,3-saturated acyl-[ACP] + NADP(+) = a (2E)-enoyl-[ACP] + H(+) + NADPH
(9Z)-octadecenoyl-[ACP] + H2O = (9Z)-octadecenoate + H(+) + holo-[ACP]
acetyl-CoA + holo-[ACP] = acetyl-[ACP] + CoA
holo-[ACP] + malonyl-CoA = CoA + malonyl-[ACP]
a fatty acyl-[ACP] + H(+) + malonyl-[ACP] = a 3-oxoacyl-[ACP] + CO2 + holo-[ACP]
a (3R)-hydroxyacyl-[ACP] + NADP(+) = a 3-oxoacyl-[ACP] + H(+) + NADPH
a (3R)-hydroxyacyl-[ACP] = a (2E)-enoyl-[ACP] + H2O
a 2,3-saturated acyl-[ACP] + NADP(+) = a (2E)-enoyl-[ACP] + H(+) + NADPH
(9Z)-octadecenoyl-[ACP] + H2O = (9Z)-octadecenoate + H(+) + holo-[ACP]
Subunit
Homodimer which is arranged in a head to tail fashion.
Keywords
Acetylation
Alternative splicing
Complete proteome
Direct protein sequencing
Fatty acid biosynthesis
Fatty acid metabolism
Hydrolase
Lipid biosynthesis
Lipid metabolism
Lyase
Multifunctional enzyme
NAD
NADP
Oxidoreductase
Phosphopantetheine
Phosphoprotein
Pyridoxal phosphate
Reference proteome
Transferase
Feature
chain Fatty acid synthase
splice variant In isoform 1.
splice variant In isoform 1.
Uniprot
O01677
H9JGF5
A0A2H1WZR1
A0A2A4IY92
A0A2A4JTZ4
A0A2H1V933
+ More
A0A3S2NJM7 A0A2A4JVR3 A0A2A4K3Y5 A0A2A4K4K9 H9JGD5 A0A2A4IZ90 A0A2H1VPQ1 A0A0N1I4Z6 A0A2H1VBX5 A0A0L7L2W8 A0A0L7L1N0 A0A2A4K3Y0 A0A194PKD7 A0A3S2NT94 A0A3S2M8J0 A0A1L8D6D5 A0A2A4K5B4 A0A1B6CGM1 A0A194QVD5 A0A1B6KCK1 A0A212EN78 A0A182YSB5 A0A182SG92 A0A182MEL3 A0A172GY34 A0A1L8D6P2 A0A1B6IHH2 A0A1B6JP24 A0A182G1R8 A0A091V5Y9 A0A182GNP7 A0A2P8XAG4 A0A182GVQ5 D2A4M7 A0A182WHF5 A0A1S4F133 Q17IW6 A0A1S3HXN4 A0A1S3I0B3 A0A087ZKN3 A0A226N0S9 U4UDB2 T1E9A6 A0A182M3X3 D2A4M5 V5GRN6 A0A182I0F0 Q7PYE4 A0A182V5F6 A0A182XBL3 A0A1W4XV87 E0W0Z3 A0A182KM38 Q16ZI9 A0A1S4FIT0 A0A226PNW4 A0A182P667 N6TW05 A0A067RHL3 A0A182JQR8 A0A1J1I2M2 A0A1D5P6Q5 A0A1S4GYC7 P12276-2 A0A182VQY2 P12276 A0A1L1RKS2 A0A3Q0IWX7 Q7Q4L2 A0A2C9GS69 A0A2J7RA25 D2A4P4 A0A093G8C4 A0A344X1T4 A0A182TPE9 A0A091ET99 A0A1S4HE27 A0A1B6GY38 A0A1J1IGX7 A0A182R1J1 N6TYU8 U4UFT3 A0A182YJT7 A0A1J1IKA4 D2A4M4 G1KJG9 A0A093S8M7 A0A182FQK1 A0A182N5X9 A0A2H1W5A9
A0A3S2NJM7 A0A2A4JVR3 A0A2A4K3Y5 A0A2A4K4K9 H9JGD5 A0A2A4IZ90 A0A2H1VPQ1 A0A0N1I4Z6 A0A2H1VBX5 A0A0L7L2W8 A0A0L7L1N0 A0A2A4K3Y0 A0A194PKD7 A0A3S2NT94 A0A3S2M8J0 A0A1L8D6D5 A0A2A4K5B4 A0A1B6CGM1 A0A194QVD5 A0A1B6KCK1 A0A212EN78 A0A182YSB5 A0A182SG92 A0A182MEL3 A0A172GY34 A0A1L8D6P2 A0A1B6IHH2 A0A1B6JP24 A0A182G1R8 A0A091V5Y9 A0A182GNP7 A0A2P8XAG4 A0A182GVQ5 D2A4M7 A0A182WHF5 A0A1S4F133 Q17IW6 A0A1S3HXN4 A0A1S3I0B3 A0A087ZKN3 A0A226N0S9 U4UDB2 T1E9A6 A0A182M3X3 D2A4M5 V5GRN6 A0A182I0F0 Q7PYE4 A0A182V5F6 A0A182XBL3 A0A1W4XV87 E0W0Z3 A0A182KM38 Q16ZI9 A0A1S4FIT0 A0A226PNW4 A0A182P667 N6TW05 A0A067RHL3 A0A182JQR8 A0A1J1I2M2 A0A1D5P6Q5 A0A1S4GYC7 P12276-2 A0A182VQY2 P12276 A0A1L1RKS2 A0A3Q0IWX7 Q7Q4L2 A0A2C9GS69 A0A2J7RA25 D2A4P4 A0A093G8C4 A0A344X1T4 A0A182TPE9 A0A091ET99 A0A1S4HE27 A0A1B6GY38 A0A1J1IGX7 A0A182R1J1 N6TYU8 U4UFT3 A0A182YJT7 A0A1J1IKA4 D2A4M4 G1KJG9 A0A093S8M7 A0A182FQK1 A0A182N5X9 A0A2H1W5A9
EC Number
2.3.1.85
Pubmed
EMBL
U67866
AAB53257.1
BABH01033637
ODYU01012325
SOQ58579.1
NWSH01005646
+ More
PCG64083.1 NWSH01000591 PCG75471.1 ODYU01001294 SOQ37311.1 RSAL01000068 RVE49259.1 PCG75472.1 NWSH01000156 PCG78967.1 PCG78966.1 BABH01033612 BABH01033613 NWSH01005017 PCG64490.1 ODYU01003708 SOQ42815.1 KQ461178 KPJ07769.1 ODYU01001484 SOQ37744.1 JTDY01003263 KOB69848.1 JTDY01003650 KOB69211.1 PCG78965.1 KQ459602 KPI93194.1 RSAL01000223 RVE44006.1 RSAL01000013 RVE53445.1 GEYN01000117 JAV02012.1 PCG78963.1 GEDC01024813 JAS12485.1 KQ461073 KPJ09498.1 GEBQ01031053 JAT08924.1 AGBW02013713 OWR42929.1 AXCM01003979 KU516006 AMK38868.1 GEYN01000115 JAV02014.1 GECU01021338 JAS86368.1 GECU01006695 JAT01012.1 JXUM01038912 KQ561184 KXJ79436.1 KL410718 KFQ98751.1 JXUM01076840 KQ562972 KXJ74732.1 PYGN01016841 PSN28987.1 JXUM01091581 JXUM01091582 KQ971344 EFA05201.1 CH477236 EAT46627.1 MCFN01000295 OXB61122.1 KB632003 ERL87905.1 GAMD01002013 JAA99577.1 AXCM01001692 EFA05203.2 GALX01004174 JAB64292.1 APCN01002072 AAAB01008987 EAA01044.4 DS235863 EEB19298.1 CH477489 EAT40085.1 AWGT02000039 OXB81109.1 APGK01052936 APGK01052937 KB741216 ENN72556.1 KK852680 KDR18643.1 CVRI01000038 CRK94451.1 AADN05000628 AAAB01008964 J03860 J04485 J02839 EAA12911.4 APCN01000761 NEVH01006580 PNF37676.1 EFA05248.1 KL215558 KFV65378.1 MF817713 AXE71619.1 KK717885 KFO52920.1 GECZ01002433 JAS67336.1 CVRI01000051 CRK99509.1 APGK01049539 APGK01049540 KB741156 ENN73526.1 KB632169 ERL89446.1 CRK99510.1 EFA05204.2 KL670490 KFW79189.1 ODYU01006431 SOQ48269.1
PCG64083.1 NWSH01000591 PCG75471.1 ODYU01001294 SOQ37311.1 RSAL01000068 RVE49259.1 PCG75472.1 NWSH01000156 PCG78967.1 PCG78966.1 BABH01033612 BABH01033613 NWSH01005017 PCG64490.1 ODYU01003708 SOQ42815.1 KQ461178 KPJ07769.1 ODYU01001484 SOQ37744.1 JTDY01003263 KOB69848.1 JTDY01003650 KOB69211.1 PCG78965.1 KQ459602 KPI93194.1 RSAL01000223 RVE44006.1 RSAL01000013 RVE53445.1 GEYN01000117 JAV02012.1 PCG78963.1 GEDC01024813 JAS12485.1 KQ461073 KPJ09498.1 GEBQ01031053 JAT08924.1 AGBW02013713 OWR42929.1 AXCM01003979 KU516006 AMK38868.1 GEYN01000115 JAV02014.1 GECU01021338 JAS86368.1 GECU01006695 JAT01012.1 JXUM01038912 KQ561184 KXJ79436.1 KL410718 KFQ98751.1 JXUM01076840 KQ562972 KXJ74732.1 PYGN01016841 PSN28987.1 JXUM01091581 JXUM01091582 KQ971344 EFA05201.1 CH477236 EAT46627.1 MCFN01000295 OXB61122.1 KB632003 ERL87905.1 GAMD01002013 JAA99577.1 AXCM01001692 EFA05203.2 GALX01004174 JAB64292.1 APCN01002072 AAAB01008987 EAA01044.4 DS235863 EEB19298.1 CH477489 EAT40085.1 AWGT02000039 OXB81109.1 APGK01052936 APGK01052937 KB741216 ENN72556.1 KK852680 KDR18643.1 CVRI01000038 CRK94451.1 AADN05000628 AAAB01008964 J03860 J04485 J02839 EAA12911.4 APCN01000761 NEVH01006580 PNF37676.1 EFA05248.1 KL215558 KFV65378.1 MF817713 AXE71619.1 KK717885 KFO52920.1 GECZ01002433 JAS67336.1 CVRI01000051 CRK99509.1 APGK01049539 APGK01049540 KB741156 ENN73526.1 KB632169 ERL89446.1 CRK99510.1 EFA05204.2 KL670490 KFW79189.1 ODYU01006431 SOQ48269.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000037510
UP000053268
+ More
UP000007151 UP000076408 UP000075901 UP000075883 UP000069940 UP000249989 UP000053283 UP000245037 UP000007266 UP000075920 UP000008820 UP000085678 UP000198323 UP000030742 UP000075840 UP000007062 UP000075903 UP000076407 UP000192223 UP000009046 UP000075882 UP000198419 UP000075885 UP000019118 UP000027135 UP000075881 UP000183832 UP000000539 UP000079169 UP000235965 UP000053875 UP000075902 UP000052976 UP000075900 UP000001646 UP000053258 UP000069272 UP000075884
UP000007151 UP000076408 UP000075901 UP000075883 UP000069940 UP000249989 UP000053283 UP000245037 UP000007266 UP000075920 UP000008820 UP000085678 UP000198323 UP000030742 UP000075840 UP000007062 UP000075903 UP000076407 UP000192223 UP000009046 UP000075882 UP000198419 UP000075885 UP000019118 UP000027135 UP000075881 UP000183832 UP000000539 UP000079169 UP000235965 UP000053875 UP000075902 UP000052976 UP000075900 UP000001646 UP000053258 UP000069272 UP000075884
PRIDE
Pfam
Interpro
IPR018201
Ketoacyl_synth_AS
+ More
IPR001031 Thioesterase
IPR014043 Acyl_transferase
IPR036291 NAD(P)-bd_dom_sf
IPR032821 KAsynt_C_assoc
IPR042104 PKS_dehydratase_sf
IPR011032 GroES-like_sf
IPR014031 Ketoacyl_synth_C
IPR023102 Fatty_acid_synthase_dom_2
IPR016035 Acyl_Trfase/lysoPLipase
IPR016036 Malonyl_transacylase_ACP-bd
IPR020807 PKS_dehydratase
IPR001227 Ac_transferase_dom_sf
IPR029058 AB_hydrolase
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR020801 PKS_acyl_transferase
IPR016039 Thiolase-like
IPR014030 Ketoacyl_synth_N
IPR020843 PKS_ER
IPR013968 PKS_KR
IPR013149 ADH_C
IPR036736 ACP-like_sf
IPR009081 PP-bd_ACP
IPR020806 PKS_PP-bd
IPR029063 SAM-dependent_MTases
IPR006162 Ppantetheine_attach_site
IPR001031 Thioesterase
IPR014043 Acyl_transferase
IPR036291 NAD(P)-bd_dom_sf
IPR032821 KAsynt_C_assoc
IPR042104 PKS_dehydratase_sf
IPR011032 GroES-like_sf
IPR014031 Ketoacyl_synth_C
IPR023102 Fatty_acid_synthase_dom_2
IPR016035 Acyl_Trfase/lysoPLipase
IPR016036 Malonyl_transacylase_ACP-bd
IPR020807 PKS_dehydratase
IPR001227 Ac_transferase_dom_sf
IPR029058 AB_hydrolase
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR020801 PKS_acyl_transferase
IPR016039 Thiolase-like
IPR014030 Ketoacyl_synth_N
IPR020843 PKS_ER
IPR013968 PKS_KR
IPR013149 ADH_C
IPR036736 ACP-like_sf
IPR009081 PP-bd_ACP
IPR020806 PKS_PP-bd
IPR029063 SAM-dependent_MTases
IPR006162 Ppantetheine_attach_site
SUPFAM
ProteinModelPortal
O01677
H9JGF5
A0A2H1WZR1
A0A2A4IY92
A0A2A4JTZ4
A0A2H1V933
+ More
A0A3S2NJM7 A0A2A4JVR3 A0A2A4K3Y5 A0A2A4K4K9 H9JGD5 A0A2A4IZ90 A0A2H1VPQ1 A0A0N1I4Z6 A0A2H1VBX5 A0A0L7L2W8 A0A0L7L1N0 A0A2A4K3Y0 A0A194PKD7 A0A3S2NT94 A0A3S2M8J0 A0A1L8D6D5 A0A2A4K5B4 A0A1B6CGM1 A0A194QVD5 A0A1B6KCK1 A0A212EN78 A0A182YSB5 A0A182SG92 A0A182MEL3 A0A172GY34 A0A1L8D6P2 A0A1B6IHH2 A0A1B6JP24 A0A182G1R8 A0A091V5Y9 A0A182GNP7 A0A2P8XAG4 A0A182GVQ5 D2A4M7 A0A182WHF5 A0A1S4F133 Q17IW6 A0A1S3HXN4 A0A1S3I0B3 A0A087ZKN3 A0A226N0S9 U4UDB2 T1E9A6 A0A182M3X3 D2A4M5 V5GRN6 A0A182I0F0 Q7PYE4 A0A182V5F6 A0A182XBL3 A0A1W4XV87 E0W0Z3 A0A182KM38 Q16ZI9 A0A1S4FIT0 A0A226PNW4 A0A182P667 N6TW05 A0A067RHL3 A0A182JQR8 A0A1J1I2M2 A0A1D5P6Q5 A0A1S4GYC7 P12276-2 A0A182VQY2 P12276 A0A1L1RKS2 A0A3Q0IWX7 Q7Q4L2 A0A2C9GS69 A0A2J7RA25 D2A4P4 A0A093G8C4 A0A344X1T4 A0A182TPE9 A0A091ET99 A0A1S4HE27 A0A1B6GY38 A0A1J1IGX7 A0A182R1J1 N6TYU8 U4UFT3 A0A182YJT7 A0A1J1IKA4 D2A4M4 G1KJG9 A0A093S8M7 A0A182FQK1 A0A182N5X9 A0A2H1W5A9
A0A3S2NJM7 A0A2A4JVR3 A0A2A4K3Y5 A0A2A4K4K9 H9JGD5 A0A2A4IZ90 A0A2H1VPQ1 A0A0N1I4Z6 A0A2H1VBX5 A0A0L7L2W8 A0A0L7L1N0 A0A2A4K3Y0 A0A194PKD7 A0A3S2NT94 A0A3S2M8J0 A0A1L8D6D5 A0A2A4K5B4 A0A1B6CGM1 A0A194QVD5 A0A1B6KCK1 A0A212EN78 A0A182YSB5 A0A182SG92 A0A182MEL3 A0A172GY34 A0A1L8D6P2 A0A1B6IHH2 A0A1B6JP24 A0A182G1R8 A0A091V5Y9 A0A182GNP7 A0A2P8XAG4 A0A182GVQ5 D2A4M7 A0A182WHF5 A0A1S4F133 Q17IW6 A0A1S3HXN4 A0A1S3I0B3 A0A087ZKN3 A0A226N0S9 U4UDB2 T1E9A6 A0A182M3X3 D2A4M5 V5GRN6 A0A182I0F0 Q7PYE4 A0A182V5F6 A0A182XBL3 A0A1W4XV87 E0W0Z3 A0A182KM38 Q16ZI9 A0A1S4FIT0 A0A226PNW4 A0A182P667 N6TW05 A0A067RHL3 A0A182JQR8 A0A1J1I2M2 A0A1D5P6Q5 A0A1S4GYC7 P12276-2 A0A182VQY2 P12276 A0A1L1RKS2 A0A3Q0IWX7 Q7Q4L2 A0A2C9GS69 A0A2J7RA25 D2A4P4 A0A093G8C4 A0A344X1T4 A0A182TPE9 A0A091ET99 A0A1S4HE27 A0A1B6GY38 A0A1J1IGX7 A0A182R1J1 N6TYU8 U4UFT3 A0A182YJT7 A0A1J1IKA4 D2A4M4 G1KJG9 A0A093S8M7 A0A182FQK1 A0A182N5X9 A0A2H1W5A9
PDB
6FN6
E-value=1.22731e-15,
Score=199
Ontologies
GO
GO:0004312
GO:0009058
GO:0016788
GO:0016491
GO:0016740
GO:0031177
GO:0016021
GO:0004316
GO:0102131
GO:0102132
GO:0004313
GO:0003960
GO:0016296
GO:0004320
GO:0004317
GO:0016295
GO:0004319
GO:0030223
GO:0008611
GO:0005886
GO:0006633
GO:0032100
GO:0006089
GO:0090557
GO:0005829
GO:0071353
GO:0042587
GO:0005623
GO:0005794
GO:0030224
GO:0047117
GO:0003697
GO:0004314
GO:0008659
GO:0004315
GO:0047451
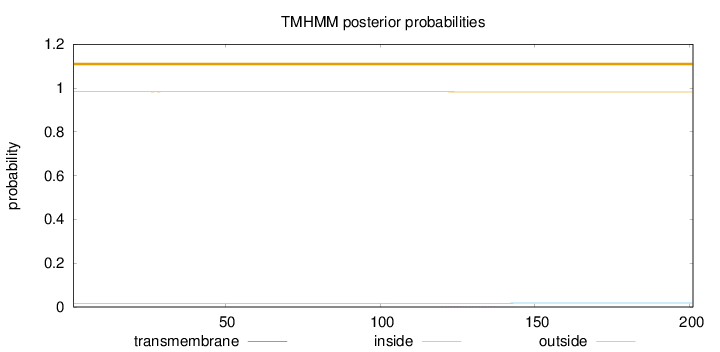
Topology
Length:
201
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01807
Exp number, first 60 AAs:
0.00251
Total prob of N-in:
0.01731
outside
1 - 201
Population Genetic Test Statistics
Pi
7.260173
Theta
15.608615
Tajima's D
-1.412545
CLR
0.722325
CSRT
0.0667966601669917
Interpretation
Uncertain
