Pre Gene Modal
BGIBMGA008239
Annotation
PREDICTED:_acyl-CoA:lysophosphatidylglycerol_acyltransferase_1-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.62 Mitochondrial Reliability : 1.429 Nuclear Reliability : 1.331
Sequence
CDS
ATGTCTAATAATGGTAAAGCGAAGAGAGATCAGTCGCTGGAGGAACTTCGGCAGCATATCCACAAATTCTACATGCCTCTCAGCAGGCAGTTCATGGTGCTCTTTCCGGAAGGTGGATTCTTGCATAAGAGAAGGGAAGTCAGTCAAAGGTTTGCCGAGAAGAATAACTTGCCGAAACTAGAATACGTATCATTACCCCGAGTTGGAGCCATGAAGGTGATCATGGAAGAAATAGGACCAAGGAGGAATGAAGGCGCTGGTACTTCTAAGGACGAATCGCGGAAAGATGACAACTTTAACCAAAGCAAACATAAGATGATAGTGGACAACAATGTGGGCGACACCATCGAATGGATCCTTGATGTGACCATCGCTTACCCTGATCGAATACCCATCCATCTTCAGGATGTCGTATGCGGGACCAGACCACCTTGCACCACACATCTTCATTACAGACTCTATCCTAGTACTGAGGTACCAACAGACACTGAGGGTATGACCCAGTGGCTATACGACAGGTTCATAGAAAAGGAGAAAATGCTGGAGGAGTTCTACAAAACCGGAAAGTTTCCCAGCAAGGGAAACCCGTCGGGATCTGTGGTCAGACAAGTACGGCAGGACAATCTTCGATACCTCATCCTGCATTCGTTCTTCATAGCTTCGACCTTCATCCAATACAAGTTCTTCAGTTTGTTGTTCAGTTATTTTTGGTAA
Protein
MSNNGKAKRDQSLEELRQHIHKFYMPLSRQFMVLFPEGGFLHKRREVSQRFAEKNNLPKLEYVSLPRVGAMKVIMEEIGPRRNEGAGTSKDESRKDDNFNQSKHKMIVDNNVGDTIEWILDVTIAYPDRIPIHLQDVVCGTRPPCTTHLHYRLYPSTEVPTDTEGMTQWLYDRFIEKEKMLEEFYKTGKFPSKGNPSGSVVRQVRQDNLRYLILHSFFIASTFIQYKFFSLLFSYFW
Summary
Similarity
Belongs to the synaptobrevin family.
Uniprot
H9JFE2
A0A2H1V948
A0A2A4JWB5
A0A1E1WC35
A0A194RIK2
A0A194QI64
+ More
A0A3S2TI19 A0A2M3ZF51 A0A2M3Z8V9 A0A2M4AJ31 A0A2M4AJK0 A0A2J7QFV8 A0A154P1D5 A0A182IRB4 A0A182VWP3 A0A151X0L8 A0A0N0U6V2 A0A195CRJ7 A0A2M4BQV9 A0A088AHM2 A0A2M4BQ02 A0A182FKW0 A0A0L7RIB0 A0A182N524 W5J7H9 B0X1Q7 D2A1V8 A0A2M4BQW0 A0A2M4CQY7 E2BRN3 F4X3K0 A0A2M4BQ05 A0A151I0U3 E9IE77 A0A2M4BQM5 A0A158NLK5 A0A195EYZ9 A0A182Q9S3 A0A1Q3FG36 A0A1Q3FG89 A0A2S2R775 A0A2S2NZX4 K7J9X8 J9JXZ3 A0A2H8TWC7 V5GRN4 A0A026W953 A0A2A3EEU1 A0A1W4X6N7 A0A1B6CCX9 A0A1B6CIT9 A0A1B6C044 A0A1Y1L3E9 E9HFY0 A0A232EY42 A0A1B6EWF5 A0A1B6INK0 A0A1B6HB95 A0A0P5M6A9 A0A146LSW6 A0A0S7GVY4 A0A1L8DYA3 A0A3Q2DQK1 A0A0S7GQU1 A0A1L8DY86 A0A1B6F3B1 W5NHT8 A0A0A9WJ04 A0A0S7GQ62 A0A3B3YE30 A0A2P8XZM5 A0A087YMF2 A0A3B3YE21 A0A3B3V176 M4ASZ4 E0VIN9 A0A3Q2Q163 A0A1W4ZE07 A0A3Q1ITG1 A0A3Q4I0J6 A0A1S3M8A4 A0A147A2T0 A0A3B1IEL1 A0A218V3M4 H0YY57 I3JFG7 A0A3Q3CVJ7 A0A3P8R0K9 A0A3B5AB86 A0A1A8NLX6 A0A1A8M0A5 A0A3P9NN15 A0A1A8GLN2 A0A1A8JCJ6 A0A1A8BEH7 A0A1A8AI30 A0A3Q3B4U6 Q4KMD5 A0A1S3JCE5
A0A3S2TI19 A0A2M3ZF51 A0A2M3Z8V9 A0A2M4AJ31 A0A2M4AJK0 A0A2J7QFV8 A0A154P1D5 A0A182IRB4 A0A182VWP3 A0A151X0L8 A0A0N0U6V2 A0A195CRJ7 A0A2M4BQV9 A0A088AHM2 A0A2M4BQ02 A0A182FKW0 A0A0L7RIB0 A0A182N524 W5J7H9 B0X1Q7 D2A1V8 A0A2M4BQW0 A0A2M4CQY7 E2BRN3 F4X3K0 A0A2M4BQ05 A0A151I0U3 E9IE77 A0A2M4BQM5 A0A158NLK5 A0A195EYZ9 A0A182Q9S3 A0A1Q3FG36 A0A1Q3FG89 A0A2S2R775 A0A2S2NZX4 K7J9X8 J9JXZ3 A0A2H8TWC7 V5GRN4 A0A026W953 A0A2A3EEU1 A0A1W4X6N7 A0A1B6CCX9 A0A1B6CIT9 A0A1B6C044 A0A1Y1L3E9 E9HFY0 A0A232EY42 A0A1B6EWF5 A0A1B6INK0 A0A1B6HB95 A0A0P5M6A9 A0A146LSW6 A0A0S7GVY4 A0A1L8DYA3 A0A3Q2DQK1 A0A0S7GQU1 A0A1L8DY86 A0A1B6F3B1 W5NHT8 A0A0A9WJ04 A0A0S7GQ62 A0A3B3YE30 A0A2P8XZM5 A0A087YMF2 A0A3B3YE21 A0A3B3V176 M4ASZ4 E0VIN9 A0A3Q2Q163 A0A1W4ZE07 A0A3Q1ITG1 A0A3Q4I0J6 A0A1S3M8A4 A0A147A2T0 A0A3B1IEL1 A0A218V3M4 H0YY57 I3JFG7 A0A3Q3CVJ7 A0A3P8R0K9 A0A3B5AB86 A0A1A8NLX6 A0A1A8M0A5 A0A3P9NN15 A0A1A8GLN2 A0A1A8JCJ6 A0A1A8BEH7 A0A1A8AI30 A0A3Q3B4U6 Q4KMD5 A0A1S3JCE5
Pubmed
EMBL
BABH01041101
ODYU01001313
SOQ37339.1
NWSH01000506
PCG75988.1
GDQN01006514
+ More
JAT84540.1 KQ460154 KPJ17269.1 KQ458880 KPJ04620.1 RSAL01000124 RVE46604.1 GGFM01006360 MBW27111.1 GGFM01004184 MBW24935.1 GGFK01007474 MBW40795.1 GGFK01007487 MBW40808.1 NEVH01014841 PNF27465.1 KQ434786 KZC05048.1 KQ982611 KYQ53897.1 KQ435727 KOX77858.1 KQ977349 KYN03368.1 GGFJ01006309 MBW55450.1 GGFJ01005902 MBW55043.1 KQ414584 KOC70569.1 ADMH02002026 ETN59916.1 DS232267 EDS38785.1 KQ971338 EFA02132.1 GGFJ01006308 MBW55449.1 GGFL01003413 MBW67591.1 GL449978 EFN81627.1 GL888616 EGI58973.1 GGFJ01006009 MBW55150.1 KQ976605 KYM79356.1 GL762562 EFZ21122.1 GGFJ01006229 MBW55370.1 ADTU01019621 KQ981914 KYN33132.1 AXCN02000175 GFDL01008527 JAV26518.1 GFDL01008482 JAV26563.1 GGMS01016347 MBY85550.1 GGMR01009993 MBY22612.1 AAZX01002316 ABLF02011377 ABLF02011378 GFXV01006414 MBW18219.1 GALX01001632 JAB66834.1 KK107378 QOIP01000012 EZA51564.1 RLU16131.1 KZ288282 PBC29531.1 GEDC01026010 JAS11288.1 GEDC01031155 GEDC01029108 GEDC01023968 JAS06143.1 JAS08190.1 JAS13330.1 GEDC01030417 JAS06881.1 GEZM01067916 GEZM01067915 JAV67328.1 GL732638 EFX69294.1 NNAY01001679 OXU23243.1 GECZ01027453 GECZ01017163 JAS42316.1 JAS52606.1 GECU01019229 GECU01014441 JAS88477.1 JAS93265.1 GECU01035786 GECU01030695 GECU01030339 GECU01028093 JAS71920.1 JAS77011.1 JAS77367.1 JAS79613.1 GDIQ01171866 GDIP01088754 LRGB01003375 JAK79859.1 JAM14961.1 KZS02990.1 GDHC01008813 JAQ09816.1 GBYX01450235 JAO31257.1 GFDF01002695 JAV11389.1 GBYX01450237 JAO31255.1 GFDF01002673 JAV11411.1 GECZ01025091 GECZ01015436 JAS44678.1 JAS54333.1 AHAT01015685 GBHO01043605 GBHO01035840 GBHO01035812 GBRD01004369 GBRD01004368 GBRD01004367 JAF99998.1 JAG07764.1 JAG07792.1 JAG61452.1 GBYX01450241 GBYX01450239 JAO31253.1 PYGN01001118 PSN37462.1 AYCK01000259 AYCK01000260 DS235201 EEB13245.1 GCES01013756 JAR72567.1 MUZQ01000055 OWK60665.1 ABQF01123287 ABQF01123288 AERX01015270 HAEI01000008 HAEH01014640 SBR69767.1 HAEF01010075 HAEG01009494 SBR49704.1 HAEB01012352 HAEC01003854 SBQ71931.1 HAED01020018 HAEE01012572 SBR06530.1 HADZ01001100 HAEA01016881 SBP65041.1 HADY01016229 HAEJ01016059 SBP54714.1 BX248403 BC098616 AAH98616.1
JAT84540.1 KQ460154 KPJ17269.1 KQ458880 KPJ04620.1 RSAL01000124 RVE46604.1 GGFM01006360 MBW27111.1 GGFM01004184 MBW24935.1 GGFK01007474 MBW40795.1 GGFK01007487 MBW40808.1 NEVH01014841 PNF27465.1 KQ434786 KZC05048.1 KQ982611 KYQ53897.1 KQ435727 KOX77858.1 KQ977349 KYN03368.1 GGFJ01006309 MBW55450.1 GGFJ01005902 MBW55043.1 KQ414584 KOC70569.1 ADMH02002026 ETN59916.1 DS232267 EDS38785.1 KQ971338 EFA02132.1 GGFJ01006308 MBW55449.1 GGFL01003413 MBW67591.1 GL449978 EFN81627.1 GL888616 EGI58973.1 GGFJ01006009 MBW55150.1 KQ976605 KYM79356.1 GL762562 EFZ21122.1 GGFJ01006229 MBW55370.1 ADTU01019621 KQ981914 KYN33132.1 AXCN02000175 GFDL01008527 JAV26518.1 GFDL01008482 JAV26563.1 GGMS01016347 MBY85550.1 GGMR01009993 MBY22612.1 AAZX01002316 ABLF02011377 ABLF02011378 GFXV01006414 MBW18219.1 GALX01001632 JAB66834.1 KK107378 QOIP01000012 EZA51564.1 RLU16131.1 KZ288282 PBC29531.1 GEDC01026010 JAS11288.1 GEDC01031155 GEDC01029108 GEDC01023968 JAS06143.1 JAS08190.1 JAS13330.1 GEDC01030417 JAS06881.1 GEZM01067916 GEZM01067915 JAV67328.1 GL732638 EFX69294.1 NNAY01001679 OXU23243.1 GECZ01027453 GECZ01017163 JAS42316.1 JAS52606.1 GECU01019229 GECU01014441 JAS88477.1 JAS93265.1 GECU01035786 GECU01030695 GECU01030339 GECU01028093 JAS71920.1 JAS77011.1 JAS77367.1 JAS79613.1 GDIQ01171866 GDIP01088754 LRGB01003375 JAK79859.1 JAM14961.1 KZS02990.1 GDHC01008813 JAQ09816.1 GBYX01450235 JAO31257.1 GFDF01002695 JAV11389.1 GBYX01450237 JAO31255.1 GFDF01002673 JAV11411.1 GECZ01025091 GECZ01015436 JAS44678.1 JAS54333.1 AHAT01015685 GBHO01043605 GBHO01035840 GBHO01035812 GBRD01004369 GBRD01004368 GBRD01004367 JAF99998.1 JAG07764.1 JAG07792.1 JAG61452.1 GBYX01450241 GBYX01450239 JAO31253.1 PYGN01001118 PSN37462.1 AYCK01000259 AYCK01000260 DS235201 EEB13245.1 GCES01013756 JAR72567.1 MUZQ01000055 OWK60665.1 ABQF01123287 ABQF01123288 AERX01015270 HAEI01000008 HAEH01014640 SBR69767.1 HAEF01010075 HAEG01009494 SBR49704.1 HAEB01012352 HAEC01003854 SBQ71931.1 HAED01020018 HAEE01012572 SBR06530.1 HADZ01001100 HAEA01016881 SBP65041.1 HADY01016229 HAEJ01016059 SBP54714.1 BX248403 BC098616 AAH98616.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000283053
UP000235965
+ More
UP000076502 UP000075880 UP000075920 UP000075809 UP000053105 UP000078542 UP000005203 UP000069272 UP000053825 UP000075884 UP000000673 UP000002320 UP000007266 UP000008237 UP000007755 UP000078540 UP000005205 UP000078541 UP000075886 UP000002358 UP000007819 UP000053097 UP000279307 UP000242457 UP000192223 UP000000305 UP000215335 UP000076858 UP000265020 UP000018468 UP000261480 UP000245037 UP000028760 UP000261500 UP000002852 UP000009046 UP000265000 UP000192224 UP000265040 UP000261580 UP000087266 UP000018467 UP000197619 UP000007754 UP000005207 UP000264840 UP000265100 UP000261400 UP000242638 UP000264800 UP000000437 UP000085678
UP000076502 UP000075880 UP000075920 UP000075809 UP000053105 UP000078542 UP000005203 UP000069272 UP000053825 UP000075884 UP000000673 UP000002320 UP000007266 UP000008237 UP000007755 UP000078540 UP000005205 UP000078541 UP000075886 UP000002358 UP000007819 UP000053097 UP000279307 UP000242457 UP000192223 UP000000305 UP000215335 UP000076858 UP000265020 UP000018468 UP000261480 UP000245037 UP000028760 UP000261500 UP000002852 UP000009046 UP000265000 UP000192224 UP000265040 UP000261580 UP000087266 UP000018467 UP000197619 UP000007754 UP000005207 UP000264840 UP000265100 UP000261400 UP000242638 UP000264800 UP000000437 UP000085678
PRIDE
Pfam
Interpro
IPR032098
Acyltransf_C
+ More
IPR002165 Plexin_repeat
IPR014756 Ig_E-set
IPR002909 IPT_dom
IPR001627 Semap_dom
IPR031148 Plexin
IPR041362 TIG2_plexin
IPR002123 Plipid/glycerol_acylTrfase
IPR013783 Ig-like_fold
IPR041019 TIG1_plexin
IPR013548 Plexin_cytoplasmic_RasGAP_dom
IPR008936 Rho_GTPase_activation_prot
IPR016201 PSI
IPR036352 Semap_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR016181 Acyl_CoA_acyltransferase
IPR000182 GNAT_dom
IPR010908 Longin_dom
IPR001388 Synaptobrevin
IPR011012 Longin-like_dom_sf
IPR002165 Plexin_repeat
IPR014756 Ig_E-set
IPR002909 IPT_dom
IPR001627 Semap_dom
IPR031148 Plexin
IPR041362 TIG2_plexin
IPR002123 Plipid/glycerol_acylTrfase
IPR013783 Ig-like_fold
IPR041019 TIG1_plexin
IPR013548 Plexin_cytoplasmic_RasGAP_dom
IPR008936 Rho_GTPase_activation_prot
IPR016201 PSI
IPR036352 Semap_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR016181 Acyl_CoA_acyltransferase
IPR000182 GNAT_dom
IPR010908 Longin_dom
IPR001388 Synaptobrevin
IPR011012 Longin-like_dom_sf
SUPFAM
Gene 3D
ProteinModelPortal
H9JFE2
A0A2H1V948
A0A2A4JWB5
A0A1E1WC35
A0A194RIK2
A0A194QI64
+ More
A0A3S2TI19 A0A2M3ZF51 A0A2M3Z8V9 A0A2M4AJ31 A0A2M4AJK0 A0A2J7QFV8 A0A154P1D5 A0A182IRB4 A0A182VWP3 A0A151X0L8 A0A0N0U6V2 A0A195CRJ7 A0A2M4BQV9 A0A088AHM2 A0A2M4BQ02 A0A182FKW0 A0A0L7RIB0 A0A182N524 W5J7H9 B0X1Q7 D2A1V8 A0A2M4BQW0 A0A2M4CQY7 E2BRN3 F4X3K0 A0A2M4BQ05 A0A151I0U3 E9IE77 A0A2M4BQM5 A0A158NLK5 A0A195EYZ9 A0A182Q9S3 A0A1Q3FG36 A0A1Q3FG89 A0A2S2R775 A0A2S2NZX4 K7J9X8 J9JXZ3 A0A2H8TWC7 V5GRN4 A0A026W953 A0A2A3EEU1 A0A1W4X6N7 A0A1B6CCX9 A0A1B6CIT9 A0A1B6C044 A0A1Y1L3E9 E9HFY0 A0A232EY42 A0A1B6EWF5 A0A1B6INK0 A0A1B6HB95 A0A0P5M6A9 A0A146LSW6 A0A0S7GVY4 A0A1L8DYA3 A0A3Q2DQK1 A0A0S7GQU1 A0A1L8DY86 A0A1B6F3B1 W5NHT8 A0A0A9WJ04 A0A0S7GQ62 A0A3B3YE30 A0A2P8XZM5 A0A087YMF2 A0A3B3YE21 A0A3B3V176 M4ASZ4 E0VIN9 A0A3Q2Q163 A0A1W4ZE07 A0A3Q1ITG1 A0A3Q4I0J6 A0A1S3M8A4 A0A147A2T0 A0A3B1IEL1 A0A218V3M4 H0YY57 I3JFG7 A0A3Q3CVJ7 A0A3P8R0K9 A0A3B5AB86 A0A1A8NLX6 A0A1A8M0A5 A0A3P9NN15 A0A1A8GLN2 A0A1A8JCJ6 A0A1A8BEH7 A0A1A8AI30 A0A3Q3B4U6 Q4KMD5 A0A1S3JCE5
A0A3S2TI19 A0A2M3ZF51 A0A2M3Z8V9 A0A2M4AJ31 A0A2M4AJK0 A0A2J7QFV8 A0A154P1D5 A0A182IRB4 A0A182VWP3 A0A151X0L8 A0A0N0U6V2 A0A195CRJ7 A0A2M4BQV9 A0A088AHM2 A0A2M4BQ02 A0A182FKW0 A0A0L7RIB0 A0A182N524 W5J7H9 B0X1Q7 D2A1V8 A0A2M4BQW0 A0A2M4CQY7 E2BRN3 F4X3K0 A0A2M4BQ05 A0A151I0U3 E9IE77 A0A2M4BQM5 A0A158NLK5 A0A195EYZ9 A0A182Q9S3 A0A1Q3FG36 A0A1Q3FG89 A0A2S2R775 A0A2S2NZX4 K7J9X8 J9JXZ3 A0A2H8TWC7 V5GRN4 A0A026W953 A0A2A3EEU1 A0A1W4X6N7 A0A1B6CCX9 A0A1B6CIT9 A0A1B6C044 A0A1Y1L3E9 E9HFY0 A0A232EY42 A0A1B6EWF5 A0A1B6INK0 A0A1B6HB95 A0A0P5M6A9 A0A146LSW6 A0A0S7GVY4 A0A1L8DYA3 A0A3Q2DQK1 A0A0S7GQU1 A0A1L8DY86 A0A1B6F3B1 W5NHT8 A0A0A9WJ04 A0A0S7GQ62 A0A3B3YE30 A0A2P8XZM5 A0A087YMF2 A0A3B3YE21 A0A3B3V176 M4ASZ4 E0VIN9 A0A3Q2Q163 A0A1W4ZE07 A0A3Q1ITG1 A0A3Q4I0J6 A0A1S3M8A4 A0A147A2T0 A0A3B1IEL1 A0A218V3M4 H0YY57 I3JFG7 A0A3Q3CVJ7 A0A3P8R0K9 A0A3B5AB86 A0A1A8NLX6 A0A1A8M0A5 A0A3P9NN15 A0A1A8GLN2 A0A1A8JCJ6 A0A1A8BEH7 A0A1A8AI30 A0A3Q3B4U6 Q4KMD5 A0A1S3JCE5
Ontologies
PANTHER
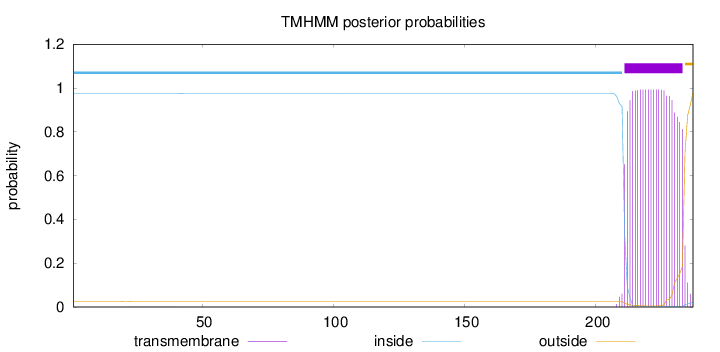
Topology
Length:
237
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.2528
Exp number, first 60 AAs:
0.01631
Total prob of N-in:
0.97410
inside
1 - 210
TMhelix
211 - 233
outside
234 - 237
Population Genetic Test Statistics
Pi
296.806628
Theta
206.012908
Tajima's D
1.464157
CLR
0.74614
CSRT
0.781060946952652
Interpretation
Uncertain
