Gene
KWMTBOMO11126
Pre Gene Modal
BGIBMGA008237
Annotation
PREDICTED:_RUN_domain-containing_protein_1_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 4.567
Sequence
CDS
ATGGAATCAAACAGCGACAATGAAAACGAAATTCCGATTTGGCAAGAACCAAGGTGTGAACCACTCGGCGCACCAAAAGAAATTTGCGATGAAAATGAATCTGAACAGAATTCAGGCGACAGATTACATGCTTTGGAAGAAGAGCAGGAGATATTATCATCCTCTGTGTTCTCCTTGACGTCTCATTTGGCACAAGTTGAGTTTCGTTTGCGACAAATTTTGAAAGCTCCTCCAGAAGAGAAAGACGCTATGCTGAAGGCTCTCGAAGAGTTCACTTCGCGTGGAGTGTCTGATACAAGAGCAGCTCCGCAAGGAAGCTCCGGTGAAGGAGCTTGTAGCGAGTGCCTCCAATTAGAACGAAAGATAAGGCGGCAACGCTCCAAGCAGACACATTTTATTGAAAGATTAAAATCACAACTGATAGAACTGGAAAGGTTCGCTGCAAATCCCGCGACTTTGAACGACGGTAAGCACAGAAATTTTATTGAACATTTAAAAAGCGAAATAGATCAAAGTCTTGAAGAAGGATGTCCATTGAGTGCTGATGAACTAAGACATCAGATCAATTGTGCTGTTCGCCAGTATGCCGACCGGCAGTTATCAAAAGATGAAATCATAACCAAACTCCAGGCTCACATAGAGGACATGCAGAAGTTTATAAAATTGATGAAAAATGAACAAACTCAAGAAAAATCTAAAACGGACCCTAAAAATGTAAAATCTGAGAGATTTGAGAAGACATTCAATAGAAATAAAAATAATGATGACCTAAGAACTGAAACCATAAACTTAATGAGAAAAGCTTCTACATTAATACAGATATTTACAGTTTCCCAGTTTGGATGTTCACCAAACACAGATAGGAGATATGAAAGAAAGTCCTCTACTATTGTCACCCATTGGGGTAATTTAAGAGCAAAGCTAGAGATGTCCATAGATGCAGTATTGCACTTAGTTTCTAGAGAGAGACCATCCCACATTATGATTGATAGTTATGACAGTGACTCTGATGAAGGAGGAATGATTATAAATGATGCACCCCTAACAACAGCTGTACGGCGACACTTAGCCATCAATCTTCGGGACTTACTACAACATGGGCTAGTGGGTCCAGAGTCGAATTCTTTAGTACCTCTAATAGGATGCTTTCCTGTACGCAGATCCTCATCCTCTCGTTGTTTGCATATATGGGATTTAATTCTTCGCTATTATGAGTTCAATGACGGTGACCACTTCAACTCCACTCCGGCTAGGAAGTTAAGTCAAAGTTTTAACTTGGATATTGTTGGAGGCAGTGCTATAACAAGTAAACAGAGTTTATTAAGTGCAATTGGAAGTATATTGGCATCACATGCTCCTTATAAAAGAAGTTATGATGCCCATTTCAAAGCCTTTGTCTGTGCAGCTTTGAATAAACATAAACTAGTTACTTGGCTGAACATAATGTTAAAATGCCGGTCTCTTGTTGATTCGTATTATTCATCAACAAGTTACATTGTAAATACTGGCTTCCAAGATGCCTTGCAAAGTCTGGATCGTTTGACTCCATACAATTTTGATTTGCCAGTAGATCTGGCTGTCAAGCAGTTTCAGAATATCAAAGATGTATTTATGTAA
Protein
MESNSDNENEIPIWQEPRCEPLGAPKEICDENESEQNSGDRLHALEEEQEILSSSVFSLTSHLAQVEFRLRQILKAPPEEKDAMLKALEEFTSRGVSDTRAAPQGSSGEGACSECLQLERKIRRQRSKQTHFIERLKSQLIELERFAANPATLNDGKHRNFIEHLKSEIDQSLEEGCPLSADELRHQINCAVRQYADRQLSKDEIITKLQAHIEDMQKFIKLMKNEQTQEKSKTDPKNVKSERFEKTFNRNKNNDDLRTETINLMRKASTLIQIFTVSQFGCSPNTDRRYERKSSTIVTHWGNLRAKLEMSIDAVLHLVSRERPSHIMIDSYDSDSDEGGMIINDAPLTTAVRRHLAINLRDLLQHGLVGPESNSLVPLIGCFPVRRSSSSRCLHIWDLILRYYEFNDGDHFNSTPARKLSQSFNLDIVGGSAITSKQSLLSAIGSILASHAPYKRSYDAHFKAFVCAALNKHKLVTWLNIMLKCRSLVDSYYSSTSYIVNTGFQDALQSLDRLTPYNFDLPVDLAVKQFQNIKDVFM
Summary
Uniprot
H9JFE0
A0A0L7KMB1
A0A2H1WPR8
A0A2A4J8G3
A0A1E1W4Y3
A0A194RIK7
+ More
A0A194QI70 A0A212FD83 A0A088A177 A0A2J7QMV4 A0A2A3EMI2 A0A067QXZ9 A0A0L7QUB6 D7EIJ8 T1J021 A0A2R5LKX0 A0A154P4X6 E2B3S1 A0A151IE87 E2ABV5 E9J4K0 A0A1B6EQD7 A0A026WGE8 A0A0C9R6Z5 A0A0C9QXP9 V5HPW8 A0A158NM66 A0A151JTW8 A0A0L8GHH4 A0A1B6CCB7 A0A0T6B425 F4WYQ0 A0A1B6MEV7 A0A0M8ZY36 A0A151WGW7 A0A151J5A7 V4AKN6 A0A023F3K7 A0A224XIN6 A0A0A9W8D7 A0A1W4XBB1 N6U857 A0A336MX84 A0A1Y1LQP1 A0A232F7Q4 A0A224Z822 A0A1E1X6Y3 A0A2R7VQB5 A0A131XJR3 A0A131YSU3 A0A023GLB8 A0A3L8DL92 T1HRR3 B7P2I2 A0A226DHI3 K1QI92 A0A2C9K3H6 A0A1B0CTY9 A0A087U782 A0A0P4W3R1 A0A2T7PIK3 E0VM57 R7TQV0 K7IQ22 A0A1V9XMK9 A0A2P8YUF6 A0A210QS98 A0A2H8U1B4 J9JSX5 A0A293LPB4 T1EFG3 A0A3Q1JGJ2 A0A3B4A360 W5N339 G3PNY2 I3JTN6 A0A3Q3F995 A0A2I4BDD2 A0A0C9Q3A0 I3JTN5 A0A3Q1FEQ3 A0A1A7X1U5 A0A3B5MSU8 A0A3B3YCP0 A0A3P9P589 A0A0F8AFQ2 A0A3Q0RNY3 A0A1A8P4P2 A0A1A8PZR9 A0A3B3UEF8 A0A3B4ZXJ0 A0A3M7PDH1 A0A3B4Y081 A0A3B4VJY1 A0A3B4GJ09 A0A3P8QVN2 A0A3P9AXT6 A0A1A8SEB3 A0A3Q3CPH0 A0A3Q1DGD2
A0A194QI70 A0A212FD83 A0A088A177 A0A2J7QMV4 A0A2A3EMI2 A0A067QXZ9 A0A0L7QUB6 D7EIJ8 T1J021 A0A2R5LKX0 A0A154P4X6 E2B3S1 A0A151IE87 E2ABV5 E9J4K0 A0A1B6EQD7 A0A026WGE8 A0A0C9R6Z5 A0A0C9QXP9 V5HPW8 A0A158NM66 A0A151JTW8 A0A0L8GHH4 A0A1B6CCB7 A0A0T6B425 F4WYQ0 A0A1B6MEV7 A0A0M8ZY36 A0A151WGW7 A0A151J5A7 V4AKN6 A0A023F3K7 A0A224XIN6 A0A0A9W8D7 A0A1W4XBB1 N6U857 A0A336MX84 A0A1Y1LQP1 A0A232F7Q4 A0A224Z822 A0A1E1X6Y3 A0A2R7VQB5 A0A131XJR3 A0A131YSU3 A0A023GLB8 A0A3L8DL92 T1HRR3 B7P2I2 A0A226DHI3 K1QI92 A0A2C9K3H6 A0A1B0CTY9 A0A087U782 A0A0P4W3R1 A0A2T7PIK3 E0VM57 R7TQV0 K7IQ22 A0A1V9XMK9 A0A2P8YUF6 A0A210QS98 A0A2H8U1B4 J9JSX5 A0A293LPB4 T1EFG3 A0A3Q1JGJ2 A0A3B4A360 W5N339 G3PNY2 I3JTN6 A0A3Q3F995 A0A2I4BDD2 A0A0C9Q3A0 I3JTN5 A0A3Q1FEQ3 A0A1A7X1U5 A0A3B5MSU8 A0A3B3YCP0 A0A3P9P589 A0A0F8AFQ2 A0A3Q0RNY3 A0A1A8P4P2 A0A1A8PZR9 A0A3B3UEF8 A0A3B4ZXJ0 A0A3M7PDH1 A0A3B4Y081 A0A3B4VJY1 A0A3B4GJ09 A0A3P8QVN2 A0A3P9AXT6 A0A1A8SEB3 A0A3Q3CPH0 A0A3Q1DGD2
Pubmed
19121390
26227816
26354079
22118469
24845553
18362917
+ More
19820115 20798317 21282665 24508170 25765539 21347285 21719571 23254933 25474469 25401762 26823975 23537049 28004739 28648823 28797301 28503490 28049606 26830274 30249741 22992520 15562597 20566863 20075255 28327890 29403074 28812685 25463417 25186727 25835551 30375419
19820115 20798317 21282665 24508170 25765539 21347285 21719571 23254933 25474469 25401762 26823975 23537049 28004739 28648823 28797301 28503490 28049606 26830274 30249741 22992520 15562597 20566863 20075255 28327890 29403074 28812685 25463417 25186727 25835551 30375419
EMBL
BABH01038326
JTDY01009131
KOB64069.1
ODYU01010153
SOQ55071.1
NWSH01002687
+ More
PCG67683.1 GDQN01009034 JAT82020.1 KQ460154 KPJ17274.1 KQ458880 KPJ04625.1 AGBW02009093 OWR51701.1 NEVH01013202 PNF29920.1 KZ288215 PBC32704.1 KK853224 KDR09703.1 KQ414735 KOC62154.1 KQ971307 EFA12041.2 JH431730 GGLE01006050 MBY10176.1 KQ434819 KZC06911.1 GL445398 EFN89658.1 KQ977890 KYM98992.1 GL438386 EFN69050.1 GL768105 EFZ12234.1 GECZ01029607 JAS40162.1 KK107238 EZA54771.1 GBYB01008572 JAG78339.1 GBYB01008564 JAG78331.1 GANP01004384 JAB80084.1 ADTU01020308 KQ981960 KYN31915.1 KQ421821 KOF76309.1 GEDC01026333 GEDC01013853 JAS10965.1 JAS23445.1 LJIG01016000 KRT81915.1 GL888455 EGI60685.1 GEBQ01005541 JAT34436.1 KQ435800 KOX73287.1 KQ983145 KYQ47062.1 KQ980039 KYN18125.1 KB201304 ESO97682.1 GBBI01003263 JAC15449.1 GFTR01006748 JAW09678.1 GBHO01040836 GBHO01037100 GBRD01009437 GDHC01021271 GDHC01007647 JAG02768.1 JAG06504.1 JAG56387.1 JAP97357.1 JAQ10982.1 APGK01039114 KB740967 KB632399 ENN76831.1 ERL94680.1 UFQS01003611 UFQT01003611 SSX15797.1 SSX35144.1 GEZM01053010 GEZM01053009 JAV74225.1 NNAY01000707 OXU26871.1 GFPF01011558 MAA22704.1 GFAC01004159 JAT95029.1 KK854027 PTY09722.1 GEFH01000787 JAP67794.1 GEDV01006233 JAP82324.1 GBBM01000747 JAC34671.1 QOIP01000007 RLU21086.1 ACPB03014367 ABJB010086659 ABJB010189896 DS622547 EEC00804.1 LNIX01000020 OXA44061.1 JH816359 EKC30909.1 AJWK01028174 KK118537 KFM73221.1 GDRN01090164 JAI60514.1 PZQS01000003 PVD33248.1 DS235290 EEB14463.1 AMQN01012797 KB309758 ELT93405.1 MNPL01007467 OQR74749.1 PYGN01000354 PSN47882.1 NEDP02002194 OWF51613.1 GFXV01008174 MBW19979.1 ABLF02036832 GFWV01004928 MAA29658.1 AMQM01004494 KB096590 ESO03552.1 AHAT01023053 AERX01008422 GBYB01008568 JAG78335.1 HADW01010591 SBP11991.1 KQ042252 KKF17042.1 HAEG01006176 SBR76044.1 HAEH01009278 SBR86736.1 REGN01011671 RMZ97039.1 HAEI01013809 SBS16278.1
PCG67683.1 GDQN01009034 JAT82020.1 KQ460154 KPJ17274.1 KQ458880 KPJ04625.1 AGBW02009093 OWR51701.1 NEVH01013202 PNF29920.1 KZ288215 PBC32704.1 KK853224 KDR09703.1 KQ414735 KOC62154.1 KQ971307 EFA12041.2 JH431730 GGLE01006050 MBY10176.1 KQ434819 KZC06911.1 GL445398 EFN89658.1 KQ977890 KYM98992.1 GL438386 EFN69050.1 GL768105 EFZ12234.1 GECZ01029607 JAS40162.1 KK107238 EZA54771.1 GBYB01008572 JAG78339.1 GBYB01008564 JAG78331.1 GANP01004384 JAB80084.1 ADTU01020308 KQ981960 KYN31915.1 KQ421821 KOF76309.1 GEDC01026333 GEDC01013853 JAS10965.1 JAS23445.1 LJIG01016000 KRT81915.1 GL888455 EGI60685.1 GEBQ01005541 JAT34436.1 KQ435800 KOX73287.1 KQ983145 KYQ47062.1 KQ980039 KYN18125.1 KB201304 ESO97682.1 GBBI01003263 JAC15449.1 GFTR01006748 JAW09678.1 GBHO01040836 GBHO01037100 GBRD01009437 GDHC01021271 GDHC01007647 JAG02768.1 JAG06504.1 JAG56387.1 JAP97357.1 JAQ10982.1 APGK01039114 KB740967 KB632399 ENN76831.1 ERL94680.1 UFQS01003611 UFQT01003611 SSX15797.1 SSX35144.1 GEZM01053010 GEZM01053009 JAV74225.1 NNAY01000707 OXU26871.1 GFPF01011558 MAA22704.1 GFAC01004159 JAT95029.1 KK854027 PTY09722.1 GEFH01000787 JAP67794.1 GEDV01006233 JAP82324.1 GBBM01000747 JAC34671.1 QOIP01000007 RLU21086.1 ACPB03014367 ABJB010086659 ABJB010189896 DS622547 EEC00804.1 LNIX01000020 OXA44061.1 JH816359 EKC30909.1 AJWK01028174 KK118537 KFM73221.1 GDRN01090164 JAI60514.1 PZQS01000003 PVD33248.1 DS235290 EEB14463.1 AMQN01012797 KB309758 ELT93405.1 MNPL01007467 OQR74749.1 PYGN01000354 PSN47882.1 NEDP02002194 OWF51613.1 GFXV01008174 MBW19979.1 ABLF02036832 GFWV01004928 MAA29658.1 AMQM01004494 KB096590 ESO03552.1 AHAT01023053 AERX01008422 GBYB01008568 JAG78335.1 HADW01010591 SBP11991.1 KQ042252 KKF17042.1 HAEG01006176 SBR76044.1 HAEH01009278 SBR86736.1 REGN01011671 RMZ97039.1 HAEI01013809 SBS16278.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000053240
UP000053268
UP000007151
+ More
UP000005203 UP000235965 UP000242457 UP000027135 UP000053825 UP000007266 UP000076502 UP000008237 UP000078542 UP000000311 UP000053097 UP000005205 UP000078541 UP000053454 UP000007755 UP000053105 UP000075809 UP000078492 UP000030746 UP000192223 UP000019118 UP000030742 UP000215335 UP000279307 UP000015103 UP000001555 UP000198287 UP000005408 UP000076420 UP000092461 UP000054359 UP000245119 UP000009046 UP000014760 UP000002358 UP000192247 UP000245037 UP000242188 UP000007819 UP000015101 UP000265040 UP000261520 UP000018468 UP000007635 UP000005207 UP000264800 UP000192220 UP000257200 UP000261380 UP000261480 UP000242638 UP000261340 UP000261500 UP000261400 UP000276133 UP000261360 UP000261420 UP000261460 UP000265100 UP000265160 UP000264840 UP000257160
UP000005203 UP000235965 UP000242457 UP000027135 UP000053825 UP000007266 UP000076502 UP000008237 UP000078542 UP000000311 UP000053097 UP000005205 UP000078541 UP000053454 UP000007755 UP000053105 UP000075809 UP000078492 UP000030746 UP000192223 UP000019118 UP000030742 UP000215335 UP000279307 UP000015103 UP000001555 UP000198287 UP000005408 UP000076420 UP000092461 UP000054359 UP000245119 UP000009046 UP000014760 UP000002358 UP000192247 UP000245037 UP000242188 UP000007819 UP000015101 UP000265040 UP000261520 UP000018468 UP000007635 UP000005207 UP000264800 UP000192220 UP000257200 UP000261380 UP000261480 UP000242638 UP000261340 UP000261500 UP000261400 UP000276133 UP000261360 UP000261420 UP000261460 UP000265100 UP000265160 UP000264840 UP000257160
Pfam
PF02759 RUN
SUPFAM
SSF140741
SSF140741
ProteinModelPortal
H9JFE0
A0A0L7KMB1
A0A2H1WPR8
A0A2A4J8G3
A0A1E1W4Y3
A0A194RIK7
+ More
A0A194QI70 A0A212FD83 A0A088A177 A0A2J7QMV4 A0A2A3EMI2 A0A067QXZ9 A0A0L7QUB6 D7EIJ8 T1J021 A0A2R5LKX0 A0A154P4X6 E2B3S1 A0A151IE87 E2ABV5 E9J4K0 A0A1B6EQD7 A0A026WGE8 A0A0C9R6Z5 A0A0C9QXP9 V5HPW8 A0A158NM66 A0A151JTW8 A0A0L8GHH4 A0A1B6CCB7 A0A0T6B425 F4WYQ0 A0A1B6MEV7 A0A0M8ZY36 A0A151WGW7 A0A151J5A7 V4AKN6 A0A023F3K7 A0A224XIN6 A0A0A9W8D7 A0A1W4XBB1 N6U857 A0A336MX84 A0A1Y1LQP1 A0A232F7Q4 A0A224Z822 A0A1E1X6Y3 A0A2R7VQB5 A0A131XJR3 A0A131YSU3 A0A023GLB8 A0A3L8DL92 T1HRR3 B7P2I2 A0A226DHI3 K1QI92 A0A2C9K3H6 A0A1B0CTY9 A0A087U782 A0A0P4W3R1 A0A2T7PIK3 E0VM57 R7TQV0 K7IQ22 A0A1V9XMK9 A0A2P8YUF6 A0A210QS98 A0A2H8U1B4 J9JSX5 A0A293LPB4 T1EFG3 A0A3Q1JGJ2 A0A3B4A360 W5N339 G3PNY2 I3JTN6 A0A3Q3F995 A0A2I4BDD2 A0A0C9Q3A0 I3JTN5 A0A3Q1FEQ3 A0A1A7X1U5 A0A3B5MSU8 A0A3B3YCP0 A0A3P9P589 A0A0F8AFQ2 A0A3Q0RNY3 A0A1A8P4P2 A0A1A8PZR9 A0A3B3UEF8 A0A3B4ZXJ0 A0A3M7PDH1 A0A3B4Y081 A0A3B4VJY1 A0A3B4GJ09 A0A3P8QVN2 A0A3P9AXT6 A0A1A8SEB3 A0A3Q3CPH0 A0A3Q1DGD2
A0A194QI70 A0A212FD83 A0A088A177 A0A2J7QMV4 A0A2A3EMI2 A0A067QXZ9 A0A0L7QUB6 D7EIJ8 T1J021 A0A2R5LKX0 A0A154P4X6 E2B3S1 A0A151IE87 E2ABV5 E9J4K0 A0A1B6EQD7 A0A026WGE8 A0A0C9R6Z5 A0A0C9QXP9 V5HPW8 A0A158NM66 A0A151JTW8 A0A0L8GHH4 A0A1B6CCB7 A0A0T6B425 F4WYQ0 A0A1B6MEV7 A0A0M8ZY36 A0A151WGW7 A0A151J5A7 V4AKN6 A0A023F3K7 A0A224XIN6 A0A0A9W8D7 A0A1W4XBB1 N6U857 A0A336MX84 A0A1Y1LQP1 A0A232F7Q4 A0A224Z822 A0A1E1X6Y3 A0A2R7VQB5 A0A131XJR3 A0A131YSU3 A0A023GLB8 A0A3L8DL92 T1HRR3 B7P2I2 A0A226DHI3 K1QI92 A0A2C9K3H6 A0A1B0CTY9 A0A087U782 A0A0P4W3R1 A0A2T7PIK3 E0VM57 R7TQV0 K7IQ22 A0A1V9XMK9 A0A2P8YUF6 A0A210QS98 A0A2H8U1B4 J9JSX5 A0A293LPB4 T1EFG3 A0A3Q1JGJ2 A0A3B4A360 W5N339 G3PNY2 I3JTN6 A0A3Q3F995 A0A2I4BDD2 A0A0C9Q3A0 I3JTN5 A0A3Q1FEQ3 A0A1A7X1U5 A0A3B5MSU8 A0A3B3YCP0 A0A3P9P589 A0A0F8AFQ2 A0A3Q0RNY3 A0A1A8P4P2 A0A1A8PZR9 A0A3B3UEF8 A0A3B4ZXJ0 A0A3M7PDH1 A0A3B4Y081 A0A3B4VJY1 A0A3B4GJ09 A0A3P8QVN2 A0A3P9AXT6 A0A1A8SEB3 A0A3Q3CPH0 A0A3Q1DGD2
Ontologies
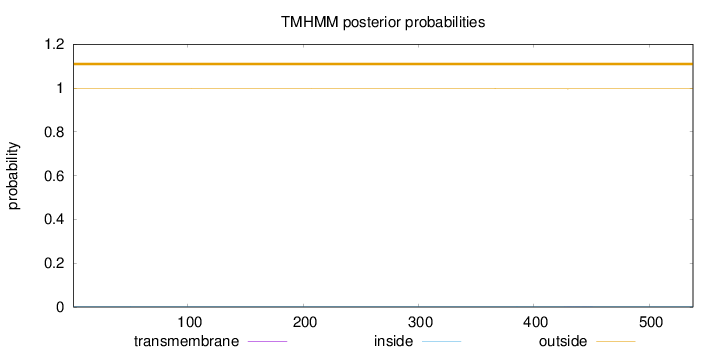
Topology
Length:
538
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02212
Exp number, first 60 AAs:
0.00029
Total prob of N-in:
0.00287
outside
1 - 538
Population Genetic Test Statistics
Pi
185.727792
Theta
213.301904
Tajima's D
-0.476433
CLR
0.645318
CSRT
0.250737463126844
Interpretation
Uncertain
