Gene
KWMTBOMO11114 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008222
Annotation
PREDICTED:_MD-2-related_lipid-recognition_protein-like_[Papilio_machaon]
Full name
MD-2-related lipid-recognition protein
Location in the cell
Extracellular Reliability : 2.63
Sequence
CDS
ATGTACGCCGTATTTTGTCTTATTTTGATTTCGAATTACGCTTTTGTTCAGGGCGAGTTTGTATTCAAGAAAAAATGTAGAGATGTGGACACTTCATTGTGTACCGTTCATAACGTGATGGTTGAACCTTGTGGCGAAGGCCCGATCTTCTGCGCTCTCAAGAAGAATAAGCCTTACAGCATAAGCTTAGACGTTACTCCACACTTTTCTGCGAATAAATTGCACGCAGTCATAAAGGGAGACGTCCAAAACCAAAACACTTTCTCGACTACCTTCACGAGATCAGCAGAGTACAATGACCTACTGGACAATACTTTATCGGAAGGGAAGCGAACACACATACAACTCCAGTTGGCTGTGGACAAACGCGCCTCTGGCAAATTCCCATTGGAAGTTCGCGTTTGGGACGAGGATGATACGTCTCATGTTTGTTGCTCAATATTTACGGTTAAAATCAGATAA
Protein
MYAVFCLILISNYAFVQGEFVFKKKCRDVDTSLCTVHNVMVEPCGEGPIFCALKKNKPYSISLDVTPHFSANKLHAVIKGDVQNQNTFSTTFTRSAEYNDLLDNTLSEGKRTHIQLQLAVDKRASGKFPLEVRVWDEDDTSHVCCSIFTVKIR
Summary
Description
Binds to lipopolysaccharide from a variety of Gram-negative bacteria and to lipid A.
Similarity
Belongs to the NPC2 family.
Keywords
Direct protein sequencing
Disulfide bond
Glycoprotein
Lipid-binding
Secreted
Signal
Feature
chain MD-2-related lipid-recognition protein
Uniprot
H9JFC5
A0A2H1VQ41
A0A2A4KA03
A0A194RI95
I4DKA2
I4DRU5
+ More
A0A212FIA0 A0A3S2LS99 A0A194QMD5 Q69FX2 I4DJ01 Q9U5E1 A0A1A9W6L0 A0A194RMQ6 A0A1B0BF04 A0A2H1VQ20 D3TR84 A0A1E1WBL4 W8BXI8 B0FHH8 A0A0M9ADU4 A0A1B0FR67 A0A195CUE1 A0A2A4K8D6 E2BVP4 A0A182QW33 A0A1A9VVQ8 A0A195BJ96 A0A026WLK4 A0A034VFC7 A0A310SNK0 Q29CB5 B4GNB9 A0A0C9REG8 F4WT68 A0A0K8W6Y5 B4LW70 A0A131YR94 A0A0L7QRS9 A0A195DEP2 A0A0A1XHF5 D2A0I8 A0A0J7L5K5 E9IGX8 A0A1W4U6E5 A0A195FNF4 B4LW71 A0A158NQ68 A0A151XFX7 A0A1Y1KP13
A0A212FIA0 A0A3S2LS99 A0A194QMD5 Q69FX2 I4DJ01 Q9U5E1 A0A1A9W6L0 A0A194RMQ6 A0A1B0BF04 A0A2H1VQ20 D3TR84 A0A1E1WBL4 W8BXI8 B0FHH8 A0A0M9ADU4 A0A1B0FR67 A0A195CUE1 A0A2A4K8D6 E2BVP4 A0A182QW33 A0A1A9VVQ8 A0A195BJ96 A0A026WLK4 A0A034VFC7 A0A310SNK0 Q29CB5 B4GNB9 A0A0C9REG8 F4WT68 A0A0K8W6Y5 B4LW70 A0A131YR94 A0A0L7QRS9 A0A195DEP2 A0A0A1XHF5 D2A0I8 A0A0J7L5K5 E9IGX8 A0A1W4U6E5 A0A195FNF4 B4LW71 A0A158NQ68 A0A151XFX7 A0A1Y1KP13
Pubmed
EMBL
BABH01038277
ODYU01003754
SOQ42918.1
NWSH01000032
PCG80482.1
KQ460154
+ More
KPJ17282.1 AK401720 KQ458880 BAM18342.1 KPJ04632.1 AK405273 BAM20635.1 AGBW02008411 OWR53463.1 RSAL01000264 RVE43288.1 KPJ04631.1 AY338398 AAQ01563.1 AK401269 BAM17891.1 AB030701 BAA89306.1 KPJ17281.1 JXJN01013235 SOQ42919.1 EZ423936 ADD20212.1 GDQN01006715 JAT84339.1 GAMC01002568 GAMC01002567 JAC03988.1 EU329722 ABY55152.1 KQ435692 KOX81109.1 CCAG010021984 KQ977279 KYN04301.1 PCG80481.1 GL450924 EFN80227.1 AXCN02001032 KQ976464 KYM84467.1 KK107154 QOIP01000001 EZA56925.1 RLU27056.1 GAKP01018709 GAKP01018708 JAC40243.1 KQ762692 OAD55535.1 CM000070 EAL26731.1 CH479186 EDW39245.1 GBYB01005361 JAG75128.1 GL888332 EGI62597.1 GDHF01005492 JAI46822.1 CH940650 EDW67604.1 GEDV01008146 JAP80411.1 KQ414771 KOC61332.1 KQ980934 KYN11307.1 GBXI01004349 JAD09943.1 KQ971338 EFA02815.1 LBMM01000477 KMQ98252.1 GL763074 EFZ20175.1 KQ981382 KYN42100.1 EDW67605.1 ADTU01022872 ADTU01022873 ADTU01022874 ADTU01022875 KQ982182 KYQ59257.1 GEZM01080352 JAV62201.1
KPJ17282.1 AK401720 KQ458880 BAM18342.1 KPJ04632.1 AK405273 BAM20635.1 AGBW02008411 OWR53463.1 RSAL01000264 RVE43288.1 KPJ04631.1 AY338398 AAQ01563.1 AK401269 BAM17891.1 AB030701 BAA89306.1 KPJ17281.1 JXJN01013235 SOQ42919.1 EZ423936 ADD20212.1 GDQN01006715 JAT84339.1 GAMC01002568 GAMC01002567 JAC03988.1 EU329722 ABY55152.1 KQ435692 KOX81109.1 CCAG010021984 KQ977279 KYN04301.1 PCG80481.1 GL450924 EFN80227.1 AXCN02001032 KQ976464 KYM84467.1 KK107154 QOIP01000001 EZA56925.1 RLU27056.1 GAKP01018709 GAKP01018708 JAC40243.1 KQ762692 OAD55535.1 CM000070 EAL26731.1 CH479186 EDW39245.1 GBYB01005361 JAG75128.1 GL888332 EGI62597.1 GDHF01005492 JAI46822.1 CH940650 EDW67604.1 GEDV01008146 JAP80411.1 KQ414771 KOC61332.1 KQ980934 KYN11307.1 GBXI01004349 JAD09943.1 KQ971338 EFA02815.1 LBMM01000477 KMQ98252.1 GL763074 EFZ20175.1 KQ981382 KYN42100.1 EDW67605.1 ADTU01022872 ADTU01022873 ADTU01022874 ADTU01022875 KQ982182 KYQ59257.1 GEZM01080352 JAV62201.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000283053
+ More
UP000091820 UP000092460 UP000053105 UP000092444 UP000078542 UP000008237 UP000075886 UP000078200 UP000078540 UP000053097 UP000279307 UP000001819 UP000008744 UP000007755 UP000008792 UP000053825 UP000078492 UP000007266 UP000036403 UP000192221 UP000078541 UP000005205 UP000075809
UP000091820 UP000092460 UP000053105 UP000092444 UP000078542 UP000008237 UP000075886 UP000078200 UP000078540 UP000053097 UP000279307 UP000001819 UP000008744 UP000007755 UP000008792 UP000053825 UP000078492 UP000007266 UP000036403 UP000192221 UP000078541 UP000005205 UP000075809
Interpro
Gene 3D
ProteinModelPortal
H9JFC5
A0A2H1VQ41
A0A2A4KA03
A0A194RI95
I4DKA2
I4DRU5
+ More
A0A212FIA0 A0A3S2LS99 A0A194QMD5 Q69FX2 I4DJ01 Q9U5E1 A0A1A9W6L0 A0A194RMQ6 A0A1B0BF04 A0A2H1VQ20 D3TR84 A0A1E1WBL4 W8BXI8 B0FHH8 A0A0M9ADU4 A0A1B0FR67 A0A195CUE1 A0A2A4K8D6 E2BVP4 A0A182QW33 A0A1A9VVQ8 A0A195BJ96 A0A026WLK4 A0A034VFC7 A0A310SNK0 Q29CB5 B4GNB9 A0A0C9REG8 F4WT68 A0A0K8W6Y5 B4LW70 A0A131YR94 A0A0L7QRS9 A0A195DEP2 A0A0A1XHF5 D2A0I8 A0A0J7L5K5 E9IGX8 A0A1W4U6E5 A0A195FNF4 B4LW71 A0A158NQ68 A0A151XFX7 A0A1Y1KP13
A0A212FIA0 A0A3S2LS99 A0A194QMD5 Q69FX2 I4DJ01 Q9U5E1 A0A1A9W6L0 A0A194RMQ6 A0A1B0BF04 A0A2H1VQ20 D3TR84 A0A1E1WBL4 W8BXI8 B0FHH8 A0A0M9ADU4 A0A1B0FR67 A0A195CUE1 A0A2A4K8D6 E2BVP4 A0A182QW33 A0A1A9VVQ8 A0A195BJ96 A0A026WLK4 A0A034VFC7 A0A310SNK0 Q29CB5 B4GNB9 A0A0C9REG8 F4WT68 A0A0K8W6Y5 B4LW70 A0A131YR94 A0A0L7QRS9 A0A195DEP2 A0A0A1XHF5 D2A0I8 A0A0J7L5K5 E9IGX8 A0A1W4U6E5 A0A195FNF4 B4LW71 A0A158NQ68 A0A151XFX7 A0A1Y1KP13
Ontologies
PANTHER
Topology
Subcellular location
Secreted
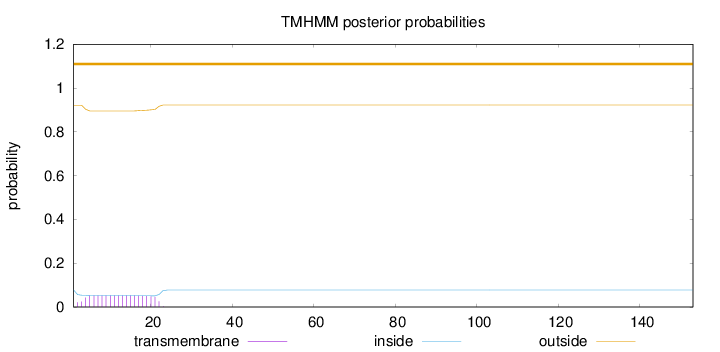
SignalP
Position: 1 - 18,
Likelihood: 0.961229

Length:
153
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.01228
Exp number, first 60 AAs:
1.01228
Total prob of N-in:
0.07832
outside
1 - 153
Population Genetic Test Statistics
Pi
342.725514
Theta
29.311906
Tajima's D
0.081883
CLR
0.082834
CSRT
0.393830308484576
Interpretation
Uncertain
