Pre Gene Modal
BGIBMGA008228
Annotation
PREDICTED:_mitochondrial_import_inner_membrane_translocase_subunit_TIM44_[Papilio_xuthus]
Full name
Mitochondrial import inner membrane translocase subunit TIM44
Location in the cell
Cytoplasmic Reliability : 1.135 Mitochondrial Reliability : 1.832 Nuclear Reliability : 1.417
Sequence
CDS
ATGCATGCATATAGACAGTTATTATATAGTCCCGCTCGTTGGAAATGTGTTCTACCTTCGAGTTTGAAGACAACATCACTCTCGGCAAAATCAGACCAGTACACCATACAAGAATGTCGGCAGTATTCTGCAAGGAAAGGATTCTTCTCATCAATAATAGAGAACATCAAAGAGGATATTGCAAAGAACAAGGAAATGAAAGAGAATATAAAGAAGTTCAGAGAGGAGGCACAGAAACTTGAGAACTCGGAAGCTTTGCAAGCAGCTAGGAAAAAGTTTCACGCCGTTGAATCAGAAGCGTCGAAGAGTTCAGAAGTCTTGAAAGAAACTATTGAGGGTATCAAAGGAAAAATGGAGCATGTGCTAGAGGAGGCGAGTAAAAGTGATATAGCCAAAAAAGCAGGGAAAATTACGGAGGACATCTCGAAGACGGCTCGCAGTGCTGCGGAGGGTATAGCGGATAAAGGACAGAGAATTGGACAAACTTCTGCTTTCAGAACTATATCGCAGGCAACAGAAGTTGTGAAAAACGAAATGTCACCCCGAGGGCTCGAGGGGCGTGTCTACATCGCTCCTGTCGCCTTAAGGAAACGTATTGAGGTCGCAGAAGACGAAAGAACGTTTTCTCCGGACACCGAGACGACTGGAGTGGAATTGCACAAGGACTCAAAGTTTTATCAGCAGTGGGAGAACTTCAAGAACAACAATCAGTATGTTAATAAGGTTTTAGATTGGAAAATCAAATACGAAGAATCGGACAATCCAGTGGTGAGGGCGTCGCGGCTGCTCACAGAAAAGATGGGAAGTCTGTTCGGGAACATGTTTGAAAAGACTGAACTATCGAAAACGTTGACGGAAATCTGTAAAATCGACCCGAACTTCACCGCACAGAAATTTCTCGAAGACTGCGCCAACGATATCATACCGAACATTCTAGAGGCTATGGTCAGAGGAGACTTGGAAATACTGAAGGACTGGTGTTACGAGGGCGTGTTCAACATTCTCGCAACCCCGATCAAGCAGTGCAAGCAGCTGGGGTATCGGCTGGACTCCAAGATCCTCGACATCGAACAGATCGAGCTGGTGATGGGCAAGATGATGGACCAGGGGCCGGTCCTCGTCATCACGTTCCAATCGCAGCAGATAATGTGTGTCAGAGACGGCAAAGGGAATGTTATCGAAGGTTCTCCGGATAAAGTTATGAGGGTTAACTACGTGTGGGTGCTGTGCCGGGACCCCCAGGAGCTCAACCCCAAGTCGGCGTGGCGGCTGCTCGAGCTGTCAGCCAACAGTGTAGAACAACTGATATAG
Protein
MHAYRQLLYSPARWKCVLPSSLKTTSLSAKSDQYTIQECRQYSARKGFFSSIIENIKEDIAKNKEMKENIKKFREEAQKLENSEALQAARKKFHAVESEASKSSEVLKETIEGIKGKMEHVLEEASKSDIAKKAGKITEDISKTARSAAEGIADKGQRIGQTSAFRTISQATEVVKNEMSPRGLEGRVYIAPVALRKRIEVAEDERTFSPDTETTGVELHKDSKFYQQWENFKNNNQYVNKVLDWKIKYEESDNPVVRASRLLTEKMGSLFGNMFEKTELSKTLTEICKIDPNFTAQKFLEDCANDIIPNILEAMVRGDLEILKDWCYEGVFNILATPIKQCKQLGYRLDSKILDIEQIELVMGKMMDQGPVLVITFQSQQIMCVRDGKGNVIEGSPDKVMRVNYVWVLCRDPQELNPKSAWRLLELSANSVEQLI
Summary
Description
Essential component of the PAM complex, a complex required for the translocation of transit peptide-containing proteins from the inner membrane into the mitochondrial matrix in an ATP-dependent manner.
Similarity
Belongs to the Tim44 family.
Uniprot
H9JFD1
A0A2A4K8T8
A0A194QGT8
A0A194RIL8
A0A3S2P6Z3
A0A2H1VKH2
+ More
A0A212EGQ1 A0A0L7LS41 A0A336LG68 A0A336LJM1 A0A023ES42 Q16YW1 Q16KX3 A0A182N1F8 A0A1B0CX04 A0A182YEY2 A0A182IQ97 A0A1L8DVE6 A0A182XBX3 A0A182QP74 A0A182MG14 A0A0K8V197 A0A1Q3F8U0 A0A182IAL1 A0A182VCZ9 A0A0A1WJA7 A0A034WTB6 A0A1J1ICR4 A0A1B6D3A0 Q7QF41 W8C6R9 A0A182RM84 A0A182TZG1 U5EHB1 W5JGB4 B0XGL1 K7J9V8 A0A182FNE5 A0A2M4AQW7 A0A182K2Y8 A0A0L0CLS3 A0A2M3Z9Y3 A0A2J7PNM9 A0A182VV88 A0A2P8YIH1 A0A084VJ49 A0A2M4BPI0 R4FJG7 A0A067QYX4 A0A182PV85 T1PIV4 A0A1I8P7L8 A0A1B6MUF4 A0A154PRU2 A0A088A218 A0A1B6I6N2 A0A1B6ID16 A0A1B6EPK7 A0A2A3EN09 A0A0A9Y9X4 A0A1A9Z8D4 A0A3B0KM66 A0A0L7QUG5 A0A151I734 A0A0M4F6I9 A0A232EJH0 A0A0M8ZZ16 A0A2R7VX32 D3TNU6 A0A0P8YHU2 A0A0Q9WZS4 B4IHY3 A0A1A9UYY1 A0A158NPU8 B3M1N4 A0A0R1E578 B4K5K9 B4LWF9 A0A1W4UPI4 A0A151X8M2 A0A0R1E0Y3 A0A1W4UBA7 A0A195ATU3 Q50LB5 B4JII9 Q9VDZ8 C9QP57 A0A0Q9WDY9 B4PMS2 A0A1A9WY78 A0A0K8TLC2 A0A1W4UMV1 A0A1W4UMN0 A0A195DTR5 A0A151JST3 Q8IHE3 Q9VDZ7 Q50LB6 A0A0R3NM32 E2BW71
A0A212EGQ1 A0A0L7LS41 A0A336LG68 A0A336LJM1 A0A023ES42 Q16YW1 Q16KX3 A0A182N1F8 A0A1B0CX04 A0A182YEY2 A0A182IQ97 A0A1L8DVE6 A0A182XBX3 A0A182QP74 A0A182MG14 A0A0K8V197 A0A1Q3F8U0 A0A182IAL1 A0A182VCZ9 A0A0A1WJA7 A0A034WTB6 A0A1J1ICR4 A0A1B6D3A0 Q7QF41 W8C6R9 A0A182RM84 A0A182TZG1 U5EHB1 W5JGB4 B0XGL1 K7J9V8 A0A182FNE5 A0A2M4AQW7 A0A182K2Y8 A0A0L0CLS3 A0A2M3Z9Y3 A0A2J7PNM9 A0A182VV88 A0A2P8YIH1 A0A084VJ49 A0A2M4BPI0 R4FJG7 A0A067QYX4 A0A182PV85 T1PIV4 A0A1I8P7L8 A0A1B6MUF4 A0A154PRU2 A0A088A218 A0A1B6I6N2 A0A1B6ID16 A0A1B6EPK7 A0A2A3EN09 A0A0A9Y9X4 A0A1A9Z8D4 A0A3B0KM66 A0A0L7QUG5 A0A151I734 A0A0M4F6I9 A0A232EJH0 A0A0M8ZZ16 A0A2R7VX32 D3TNU6 A0A0P8YHU2 A0A0Q9WZS4 B4IHY3 A0A1A9UYY1 A0A158NPU8 B3M1N4 A0A0R1E578 B4K5K9 B4LWF9 A0A1W4UPI4 A0A151X8M2 A0A0R1E0Y3 A0A1W4UBA7 A0A195ATU3 Q50LB5 B4JII9 Q9VDZ8 C9QP57 A0A0Q9WDY9 B4PMS2 A0A1A9WY78 A0A0K8TLC2 A0A1W4UMV1 A0A1W4UMN0 A0A195DTR5 A0A151JST3 Q8IHE3 Q9VDZ7 Q50LB6 A0A0R3NM32 E2BW71
Pubmed
19121390
26354079
22118469
26227816
24945155
26483478
+ More
17510324 25244985 25830018 25348373 12364791 14747013 17210077 24495485 20920257 23761445 20075255 26108605 29403074 24438588 24845553 25315136 25401762 26823975 28648823 20353571 17994087 21347285 17550304 16143626 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26369729 15632085 20798317
17510324 25244985 25830018 25348373 12364791 14747013 17210077 24495485 20920257 23761445 20075255 26108605 29403074 24438588 24845553 25315136 25401762 26823975 28648823 20353571 17994087 21347285 17550304 16143626 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26369729 15632085 20798317
EMBL
BABH01038264
BABH01038265
NWSH01000032
PCG80489.1
KQ458880
KPJ04634.1
+ More
KQ460154 KPJ17284.1 RSAL01000264 RVE43290.1 ODYU01002804 SOQ40754.1 AGBW02015051 OWR40656.1 JTDY01000262 KOB78021.1 UFQS01003369 UFQT01003369 SSX15565.1 SSX34929.1 UFQS01005995 UFQT01004579 UFQT01005995 SSX16903.1 SSX35722.1 JXUM01012907 JXUM01093602 GAPW01001543 GAPW01001542 KQ564068 KQ560364 JAC12055.1 KXJ72843.1 KXJ82804.1 CH477507 EAT39827.1 CH477933 EAT34960.1 AJWK01033084 GFDF01003670 JAV10414.1 AXCN02001377 AXCM01003602 GDHF01019964 JAI32350.1 GFDL01011112 JAV23933.1 APCN01001262 GBXI01015814 JAC98477.1 GAKP01000181 JAC58771.1 CVRI01000044 CRK96766.1 GEDC01017243 JAS20055.1 AAAB01008846 EAA06726.4 GAMC01000967 JAC05589.1 GANO01003100 JAB56771.1 ADMH02001484 ETN62378.1 DS233038 EDS27675.1 AAZX01002793 GGFK01009791 MBW43112.1 JRES01000228 KNC33187.1 GGFM01004534 MBW25285.1 NEVH01023380 PNF17947.1 PYGN01000570 PSN44050.1 ATLV01013431 KE524855 KFB37993.1 GGFJ01005557 MBW54698.1 ACPB03008137 GAHY01002283 JAA75227.1 KK852867 KDR14720.1 KA647853 AFP62482.1 GEBQ01000401 JAT39576.1 KQ435078 KZC14457.1 GECU01025110 JAS82596.1 GECU01022889 JAS84817.1 GECZ01029903 JAS39866.1 KZ288215 PBC32586.1 GBHO01045164 GBHO01015188 GBRD01013649 GDHC01011471 GDHC01003494 JAF98439.1 JAG28416.1 JAG52177.1 JAQ07158.1 JAQ15135.1 OUUW01000013 SPP87669.1 KQ414735 KOC62302.1 KQ978456 KYM93898.1 CP012526 ALC47675.1 NNAY01004007 OXU18506.1 KQ435821 KOX72479.1 KK854110 PTY11461.1 CCAG010012055 EZ423098 ADD19374.1 CH902617 KPU80814.1 CH933806 KRG00825.1 CH480840 EDW49500.1 ADTU01022755 EDV44364.1 CM000160 KRK02826.1 EDW14046.1 CH940650 EDW66596.1 KQ982409 KYQ56726.1 KRK02825.1 KQ976745 KYM75477.1 AB194415 BAD98173.1 CH916369 EDV93070.1 AE014297 BT133180 AAF55640.1 AFA28421.1 BT099939 ACX36515.1 KRF82824.1 EDW95992.1 GDAI01002625 JAI14978.1 KQ980487 KYN15919.1 KQ982014 KYN30632.1 BT001289 BT011399 AAN71045.1 AAR96191.1 AAF55639.1 BAD98172.1 CM000070 KRS99925.1 GL451091 EFN80103.1
KQ460154 KPJ17284.1 RSAL01000264 RVE43290.1 ODYU01002804 SOQ40754.1 AGBW02015051 OWR40656.1 JTDY01000262 KOB78021.1 UFQS01003369 UFQT01003369 SSX15565.1 SSX34929.1 UFQS01005995 UFQT01004579 UFQT01005995 SSX16903.1 SSX35722.1 JXUM01012907 JXUM01093602 GAPW01001543 GAPW01001542 KQ564068 KQ560364 JAC12055.1 KXJ72843.1 KXJ82804.1 CH477507 EAT39827.1 CH477933 EAT34960.1 AJWK01033084 GFDF01003670 JAV10414.1 AXCN02001377 AXCM01003602 GDHF01019964 JAI32350.1 GFDL01011112 JAV23933.1 APCN01001262 GBXI01015814 JAC98477.1 GAKP01000181 JAC58771.1 CVRI01000044 CRK96766.1 GEDC01017243 JAS20055.1 AAAB01008846 EAA06726.4 GAMC01000967 JAC05589.1 GANO01003100 JAB56771.1 ADMH02001484 ETN62378.1 DS233038 EDS27675.1 AAZX01002793 GGFK01009791 MBW43112.1 JRES01000228 KNC33187.1 GGFM01004534 MBW25285.1 NEVH01023380 PNF17947.1 PYGN01000570 PSN44050.1 ATLV01013431 KE524855 KFB37993.1 GGFJ01005557 MBW54698.1 ACPB03008137 GAHY01002283 JAA75227.1 KK852867 KDR14720.1 KA647853 AFP62482.1 GEBQ01000401 JAT39576.1 KQ435078 KZC14457.1 GECU01025110 JAS82596.1 GECU01022889 JAS84817.1 GECZ01029903 JAS39866.1 KZ288215 PBC32586.1 GBHO01045164 GBHO01015188 GBRD01013649 GDHC01011471 GDHC01003494 JAF98439.1 JAG28416.1 JAG52177.1 JAQ07158.1 JAQ15135.1 OUUW01000013 SPP87669.1 KQ414735 KOC62302.1 KQ978456 KYM93898.1 CP012526 ALC47675.1 NNAY01004007 OXU18506.1 KQ435821 KOX72479.1 KK854110 PTY11461.1 CCAG010012055 EZ423098 ADD19374.1 CH902617 KPU80814.1 CH933806 KRG00825.1 CH480840 EDW49500.1 ADTU01022755 EDV44364.1 CM000160 KRK02826.1 EDW14046.1 CH940650 EDW66596.1 KQ982409 KYQ56726.1 KRK02825.1 KQ976745 KYM75477.1 AB194415 BAD98173.1 CH916369 EDV93070.1 AE014297 BT133180 AAF55640.1 AFA28421.1 BT099939 ACX36515.1 KRF82824.1 EDW95992.1 GDAI01002625 JAI14978.1 KQ980487 KYN15919.1 KQ982014 KYN30632.1 BT001289 BT011399 AAN71045.1 AAR96191.1 AAF55639.1 BAD98172.1 CM000070 KRS99925.1 GL451091 EFN80103.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000283053
UP000007151
+ More
UP000037510 UP000069940 UP000249989 UP000008820 UP000075884 UP000092461 UP000076408 UP000075880 UP000076407 UP000075886 UP000075883 UP000075840 UP000075903 UP000183832 UP000007062 UP000075900 UP000075902 UP000000673 UP000002320 UP000002358 UP000069272 UP000075881 UP000037069 UP000235965 UP000075920 UP000245037 UP000030765 UP000015103 UP000027135 UP000075885 UP000095301 UP000095300 UP000076502 UP000005203 UP000242457 UP000092445 UP000268350 UP000053825 UP000078542 UP000092553 UP000215335 UP000053105 UP000092444 UP000007801 UP000009192 UP000001292 UP000078200 UP000005205 UP000002282 UP000008792 UP000192221 UP000075809 UP000078540 UP000001070 UP000000803 UP000091820 UP000078492 UP000078541 UP000001819 UP000008237
UP000037510 UP000069940 UP000249989 UP000008820 UP000075884 UP000092461 UP000076408 UP000075880 UP000076407 UP000075886 UP000075883 UP000075840 UP000075903 UP000183832 UP000007062 UP000075900 UP000075902 UP000000673 UP000002320 UP000002358 UP000069272 UP000075881 UP000037069 UP000235965 UP000075920 UP000245037 UP000030765 UP000015103 UP000027135 UP000075885 UP000095301 UP000095300 UP000076502 UP000005203 UP000242457 UP000092445 UP000268350 UP000053825 UP000078542 UP000092553 UP000215335 UP000053105 UP000092444 UP000007801 UP000009192 UP000001292 UP000078200 UP000005205 UP000002282 UP000008792 UP000192221 UP000075809 UP000078540 UP000001070 UP000000803 UP000091820 UP000078492 UP000078541 UP000001819 UP000008237
Interpro
Gene 3D
ProteinModelPortal
H9JFD1
A0A2A4K8T8
A0A194QGT8
A0A194RIL8
A0A3S2P6Z3
A0A2H1VKH2
+ More
A0A212EGQ1 A0A0L7LS41 A0A336LG68 A0A336LJM1 A0A023ES42 Q16YW1 Q16KX3 A0A182N1F8 A0A1B0CX04 A0A182YEY2 A0A182IQ97 A0A1L8DVE6 A0A182XBX3 A0A182QP74 A0A182MG14 A0A0K8V197 A0A1Q3F8U0 A0A182IAL1 A0A182VCZ9 A0A0A1WJA7 A0A034WTB6 A0A1J1ICR4 A0A1B6D3A0 Q7QF41 W8C6R9 A0A182RM84 A0A182TZG1 U5EHB1 W5JGB4 B0XGL1 K7J9V8 A0A182FNE5 A0A2M4AQW7 A0A182K2Y8 A0A0L0CLS3 A0A2M3Z9Y3 A0A2J7PNM9 A0A182VV88 A0A2P8YIH1 A0A084VJ49 A0A2M4BPI0 R4FJG7 A0A067QYX4 A0A182PV85 T1PIV4 A0A1I8P7L8 A0A1B6MUF4 A0A154PRU2 A0A088A218 A0A1B6I6N2 A0A1B6ID16 A0A1B6EPK7 A0A2A3EN09 A0A0A9Y9X4 A0A1A9Z8D4 A0A3B0KM66 A0A0L7QUG5 A0A151I734 A0A0M4F6I9 A0A232EJH0 A0A0M8ZZ16 A0A2R7VX32 D3TNU6 A0A0P8YHU2 A0A0Q9WZS4 B4IHY3 A0A1A9UYY1 A0A158NPU8 B3M1N4 A0A0R1E578 B4K5K9 B4LWF9 A0A1W4UPI4 A0A151X8M2 A0A0R1E0Y3 A0A1W4UBA7 A0A195ATU3 Q50LB5 B4JII9 Q9VDZ8 C9QP57 A0A0Q9WDY9 B4PMS2 A0A1A9WY78 A0A0K8TLC2 A0A1W4UMV1 A0A1W4UMN0 A0A195DTR5 A0A151JST3 Q8IHE3 Q9VDZ7 Q50LB6 A0A0R3NM32 E2BW71
A0A212EGQ1 A0A0L7LS41 A0A336LG68 A0A336LJM1 A0A023ES42 Q16YW1 Q16KX3 A0A182N1F8 A0A1B0CX04 A0A182YEY2 A0A182IQ97 A0A1L8DVE6 A0A182XBX3 A0A182QP74 A0A182MG14 A0A0K8V197 A0A1Q3F8U0 A0A182IAL1 A0A182VCZ9 A0A0A1WJA7 A0A034WTB6 A0A1J1ICR4 A0A1B6D3A0 Q7QF41 W8C6R9 A0A182RM84 A0A182TZG1 U5EHB1 W5JGB4 B0XGL1 K7J9V8 A0A182FNE5 A0A2M4AQW7 A0A182K2Y8 A0A0L0CLS3 A0A2M3Z9Y3 A0A2J7PNM9 A0A182VV88 A0A2P8YIH1 A0A084VJ49 A0A2M4BPI0 R4FJG7 A0A067QYX4 A0A182PV85 T1PIV4 A0A1I8P7L8 A0A1B6MUF4 A0A154PRU2 A0A088A218 A0A1B6I6N2 A0A1B6ID16 A0A1B6EPK7 A0A2A3EN09 A0A0A9Y9X4 A0A1A9Z8D4 A0A3B0KM66 A0A0L7QUG5 A0A151I734 A0A0M4F6I9 A0A232EJH0 A0A0M8ZZ16 A0A2R7VX32 D3TNU6 A0A0P8YHU2 A0A0Q9WZS4 B4IHY3 A0A1A9UYY1 A0A158NPU8 B3M1N4 A0A0R1E578 B4K5K9 B4LWF9 A0A1W4UPI4 A0A151X8M2 A0A0R1E0Y3 A0A1W4UBA7 A0A195ATU3 Q50LB5 B4JII9 Q9VDZ8 C9QP57 A0A0Q9WDY9 B4PMS2 A0A1A9WY78 A0A0K8TLC2 A0A1W4UMV1 A0A1W4UMN0 A0A195DTR5 A0A151JST3 Q8IHE3 Q9VDZ7 Q50LB6 A0A0R3NM32 E2BW71
PDB
2CW9
E-value=7.5224e-65,
Score=628
Ontologies
GO
PANTHER
Topology
Subcellular location
Mitochondrion inner membrane
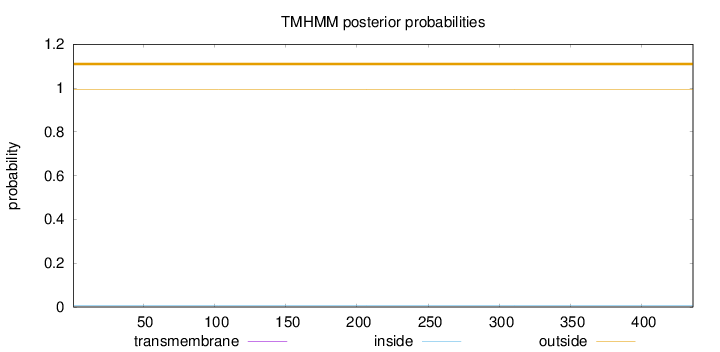
Length:
436
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00019
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00594
outside
1 - 436
Population Genetic Test Statistics
Pi
181.546129
Theta
17.352915
Tajima's D
0.929384
CLR
0.289948
CSRT
0.649317534123294
Interpretation
Uncertain
