Gene
KWMTBOMO11110 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008226
Annotation
eukaryotic_translation_initiation_factor_5_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.628
Sequence
CDS
ATGGGTTCCTTAAACGTGAACCGTAACGTCTCCGATGCGTTCTACCGGTACAAGATGCCGCGGATATGCGCCAAGGTCGAAGGGAAGGGGAACGGCATCAAGACGGTCATCGTCAACATGCCTGAAGTCGCCAAGGCGCTGGGGAGACCAGCCACGTACCCAACAAAGTACTTTGGTTGTGAATTGGGTGCCCAGACTCAGTTTGACTTCAAGAATGAGCGTTTCATTGTCAACGGAAGCCATGACTCATCTAAACTTCAAGATCTATTGGACGGATTTATTCGTAAATTCGTGCTGTGCCCTGAGTGCGACAACCCCGAGACCGAGCTGATTGTCTCCACGAAGCGGAACACCATCTCCCAAGGATGCAAGGCCTGCGGTTACCATGGACAGCTTGACTTCAATCACAAGTTGAATACTTTTATACTCAAGAACCCGCCTGCCAATGATCCGGCTGTGCAAGGCTCTTCACTGACTGAGGGCAATCGTGGAAAACGCTCGAAGCGCTCTGGTCCTGGATCCAACAACACTGCGACCAACGGCAGCCACGAGGGACAAGAAACTGAAGCTAAAAATGAACCGGAAGCGGTGTCGAGTCCTACAGCTACCGCAGCTCCGAGCACTAAGAAGGAGAAGTCCAAGAAAACTGGCGATGATGGCGATGAGGACGACGACGGCTGGACCGTGGATGTCAGCGAGGAGGCTGTGCGGGCCAGGATGCAAGATCTAACTGAAGGAGCTAAGATCATAACATTGTCAGAAGACAGTGAGAAGAATGAGAAGCAGCGGATGGATCTGTTCTATGCATTTTTGAAGCAACGAGCAGCCAGCGGTCAGGTGCAGGGAACCAAAGCCATCTCTGAGATCCTGCACGAGGCCGAGAGATTGGAAGTGAAGTCGAAGGCGCCGCTGGTGCTGTTCGAGGTGCTGGTGAACGCGGCGTCGGTGGGCGGCGACGTGAAGAAGCACCGCGCTCTGCTGCTGCGCTTCACGCGCCAAGACCCGCGCGCGCAGCGAGCCGCCCTGCACGCCCTCACCGCGCTCGCGCACCACCACGCCGCGCTGCTGCCGCGCGTGCCCGCCGTGCTCAAGCTTCTTTACGACGAAGACATCGTAGAAGAGAAGGCTATACTGGAGTGGGCTAGCAAACCATCGCGCAAATACGCATCCAAGGAAGTCGTAGCCGAAGTGAGGAAACGGGCCCAACCCTTCCTCGACTGGCTGCAGCAGGCCGAGGAGGACTCGAGTGACGACGACGACGAGGAGGAAGATATCGAGATCGAGTACGACGACCGCGCCAAGGCGACCCCCATCAAGGCCGTGGCGACTGCGGCCAACAACAAACCGAAGCAGGAAGAAGACGATGACGATTTCGATATCGACGCCATTTAA
Protein
MGSLNVNRNVSDAFYRYKMPRICAKVEGKGNGIKTVIVNMPEVAKALGRPATYPTKYFGCELGAQTQFDFKNERFIVNGSHDSSKLQDLLDGFIRKFVLCPECDNPETELIVSTKRNTISQGCKACGYHGQLDFNHKLNTFILKNPPANDPAVQGSSLTEGNRGKRSKRSGPGSNNTATNGSHEGQETEAKNEPEAVSSPTATAAPSTKKEKSKKTGDDGDEDDDGWTVDVSEEAVRARMQDLTEGAKIITLSEDSEKNEKQRMDLFYAFLKQRAASGQVQGTKAISEILHEAERLEVKSKAPLVLFEVLVNAASVGGDVKKHRALLLRFTRQDPRAQRAALHALTALAHHHAALLPRVPAVLKLLYDEDIVEEKAILEWASKPSRKYASKEVVAEVRKRAQPFLDWLQQAEEDSSDDDDEEEDIEIEYDDRAKATPIKAVATAANNKPKQEEDDDDFDIDAI
Summary
Uniprot
Q0ZB74
A0A2H1VIU9
A0A1E1W4Z3
A0A194QME0
A0A194RIA0
A0A212EGP5
+ More
Q1A4T6 A0A2A4K9U5 S5N3Z2 S4PFZ8 A0A1B6MII3 A0A0K8TRP3 A0A0C9Q139 T1D408 A0A2J7PWA2 A0A026VWF8 A0A0P4VTX6 R4FNT1 A0A2A3ERU2 A0A088AP20 A0A1B6E3K8 A0A224XNV5 A0A067QT43 T1E2M2 K7J337 A0A0M8ZT33 E2ATE1 A0A0A9X1L2 A0A069DTN5 A0A154PNS0 A0A023F8Y9 A0A195D5F3 A0A1W4X3Q3 A0A195EJS1 A0A195BFU0 F4X2B7 A0A151X6D1 A0A195FN48 A0A158NJN0 A0A1J1HPP9 U5EXY4 T1DPV6 A0A0T6AZX0 W5J9Y9 D2A3J2 A0A2M4BM37 A0A2M4A2P3 A0A2M4BM36 A0A1L8DSB3 A0A2M3YZ48 A0A084W3J1 N6UGW8 A0A182FVI3 A0A2M3YZ22 A0A0L7QRG6 A0A1I8PQZ4 A0A182PI91 A0A182G5X2 A0A182J216 A0A336JZ43 A0A182MXI5 A0A182YEB8 A0A023ESH2 A0A0L0BM06 A0A182TDI3 A0A182RCZ2 A0A182WDE1 A0A182ME45 A0A1I8JW08 A0A182VL43 A0A182LHN7 Q7QHJ0 A0A182HFX9 Q17PW1 T1PCU9 A0A2H8TV07 A0A1Q3F8L5 A0A182KC71 A0A182QXD1 B0W0M0 A0A1Y1L6V3 J9K088 A0A0J7L661 E0VLG6 W8B8V3 A0A1A9WSZ9 A0A1A9X8Q0 A0A1B0C0E7 A0A034VGT6 A0A0A1XFT7 A0A1A9ZWY2 A0A0K8SM34 A0A0K8VE40 A0A1A9UQE2 B4L5G0 A0A3Q0JG87 B4NCA3 B4MAX1 A0A1B0CSU2 A0A3B0KKX5
Q1A4T6 A0A2A4K9U5 S5N3Z2 S4PFZ8 A0A1B6MII3 A0A0K8TRP3 A0A0C9Q139 T1D408 A0A2J7PWA2 A0A026VWF8 A0A0P4VTX6 R4FNT1 A0A2A3ERU2 A0A088AP20 A0A1B6E3K8 A0A224XNV5 A0A067QT43 T1E2M2 K7J337 A0A0M8ZT33 E2ATE1 A0A0A9X1L2 A0A069DTN5 A0A154PNS0 A0A023F8Y9 A0A195D5F3 A0A1W4X3Q3 A0A195EJS1 A0A195BFU0 F4X2B7 A0A151X6D1 A0A195FN48 A0A158NJN0 A0A1J1HPP9 U5EXY4 T1DPV6 A0A0T6AZX0 W5J9Y9 D2A3J2 A0A2M4BM37 A0A2M4A2P3 A0A2M4BM36 A0A1L8DSB3 A0A2M3YZ48 A0A084W3J1 N6UGW8 A0A182FVI3 A0A2M3YZ22 A0A0L7QRG6 A0A1I8PQZ4 A0A182PI91 A0A182G5X2 A0A182J216 A0A336JZ43 A0A182MXI5 A0A182YEB8 A0A023ESH2 A0A0L0BM06 A0A182TDI3 A0A182RCZ2 A0A182WDE1 A0A182ME45 A0A1I8JW08 A0A182VL43 A0A182LHN7 Q7QHJ0 A0A182HFX9 Q17PW1 T1PCU9 A0A2H8TV07 A0A1Q3F8L5 A0A182KC71 A0A182QXD1 B0W0M0 A0A1Y1L6V3 J9K088 A0A0J7L661 E0VLG6 W8B8V3 A0A1A9WSZ9 A0A1A9X8Q0 A0A1B0C0E7 A0A034VGT6 A0A0A1XFT7 A0A1A9ZWY2 A0A0K8SM34 A0A0K8VE40 A0A1A9UQE2 B4L5G0 A0A3Q0JG87 B4NCA3 B4MAX1 A0A1B0CSU2 A0A3B0KKX5
Pubmed
26354079
22118469
16760394
23861954
23622113
26369729
+ More
24330624 24508170 30249741 27129103 24845553 20075255 20798317 25401762 26823975 26334808 25474469 21719571 21347285 20920257 23761445 18362917 19820115 24438588 23537049 26483478 25244985 24945155 26108605 20966253 12364791 14747013 17210077 17510324 25315136 28004739 20566863 24495485 25348373 25830018 17994087
24330624 24508170 30249741 27129103 24845553 20075255 20798317 25401762 26823975 26334808 25474469 21719571 21347285 20920257 23761445 18362917 19820115 24438588 23537049 26483478 25244985 24945155 26108605 20966253 12364791 14747013 17210077 17510324 25315136 28004739 20566863 24495485 25348373 25830018 17994087
EMBL
DQ645462
ABG54290.1
ODYU01002804
SOQ40753.1
GDQN01009007
GDQN01000841
+ More
GDQN01000314 JAT82047.1 JAT90213.1 JAT90740.1 KQ458880 KPJ04636.1 KQ460154 KPJ17287.1 AGBW02015051 OWR40655.1 DQ333351 ABC61144.1 NWSH01000032 PCG80490.1 KC961304 AGR57075.1 GAIX01003832 JAA88728.1 GEBQ01019059 GEBQ01016528 GEBQ01004282 JAT20918.1 JAT23449.1 JAT35695.1 GDAI01000569 JAI17034.1 GBYB01007628 JAG77395.1 GALA01001148 JAA93704.1 NEVH01020936 PNF20601.1 KK107928 QOIP01000001 EZA47159.1 RLU27049.1 GDKW01000497 JAI56098.1 ACPB03017681 GAHY01000453 JAA77057.1 KZ288193 PBC33992.1 GEDC01024965 GEDC01013149 GEDC01004819 JAS12333.1 JAS24149.1 JAS32479.1 GFTR01006306 JAW10120.1 KK852969 KDR13109.1 GALA01001145 JAA93707.1 KQ435859 KOX70570.1 GL442548 EFN63308.1 GBHO01030053 GBHO01030052 GBRD01011961 GBRD01011960 GDHC01014561 GDHC01008306 JAG13551.1 JAG13552.1 JAG53863.1 JAQ04068.1 JAQ10323.1 GBGD01001456 JAC87433.1 KQ435007 KZC13531.1 GBBI01000791 JAC17921.1 KQ976818 KYN08113.1 KQ978782 KYN28391.1 KQ976490 KYM83468.1 GL888575 EGI59399.1 KQ982482 KYQ55946.1 KQ981424 KYN41821.1 ADTU01018078 CVRI01000015 CRK90032.1 GANO01000705 JAB59166.1 GAMD01002999 JAA98591.1 LJIG01016388 KRT80751.1 ADMH02002067 ETN59670.1 KQ971338 EFA01900.1 GGFJ01005008 MBW54149.1 GGFK01001742 MBW35063.1 GGFJ01005009 MBW54150.1 GFDF01004793 JAV09291.1 GGFM01000737 MBW21488.1 ATLV01020054 KE525293 KFB44785.1 APGK01008952 APGK01020978 APGK01051622 KB741197 KB740274 KB737822 KB632308 ENN72983.1 ENN80970.1 ENN82908.1 ERL91965.1 GGFM01000762 MBW21513.1 KQ414778 KOC61218.1 JXUM01044847 KQ561415 KXJ78656.1 UFQS01000027 UFQT01000027 SSW97719.1 SSX18105.1 GAPW01001401 JAC12197.1 JRES01001651 KNC21155.1 AXCM01008583 AAAB01008816 EAA05225.2 APCN01005436 CH477188 EAT48834.1 KA646479 AFP61108.1 GFXV01006290 MBW18095.1 GFDL01011125 JAV23920.1 AXCN02001714 DS231818 EDS41322.1 GEZM01063108 GEZM01063107 JAV69304.1 ABLF02031739 ABLF02031741 LBMM01000532 KMQ98121.1 DS235271 EEB14222.1 GAMC01016950 GAMC01016949 JAB89606.1 JXJN01023561 GAKP01017268 GAKP01017267 JAC41684.1 GBXI01004496 JAD09796.1 GBRD01017772 GBRD01011959 JAG53865.1 GDHF01031099 GDHF01028759 GDHF01028251 GDHF01022750 GDHF01022203 GDHF01017863 GDHF01015529 GDHF01012742 GDHF01008220 GDHF01006614 GDHF01005950 JAI21215.1 JAI23555.1 JAI24063.1 JAI29564.1 JAI30111.1 JAI34451.1 JAI36785.1 JAI39572.1 JAI44094.1 JAI45700.1 JAI46364.1 CH933811 EDW06419.1 CH964239 EDW82462.1 CH940655 EDW66380.1 AJWK01026604 AJWK01026605 AJWK01026606 OUUW01000011 SPP86446.1
GDQN01000314 JAT82047.1 JAT90213.1 JAT90740.1 KQ458880 KPJ04636.1 KQ460154 KPJ17287.1 AGBW02015051 OWR40655.1 DQ333351 ABC61144.1 NWSH01000032 PCG80490.1 KC961304 AGR57075.1 GAIX01003832 JAA88728.1 GEBQ01019059 GEBQ01016528 GEBQ01004282 JAT20918.1 JAT23449.1 JAT35695.1 GDAI01000569 JAI17034.1 GBYB01007628 JAG77395.1 GALA01001148 JAA93704.1 NEVH01020936 PNF20601.1 KK107928 QOIP01000001 EZA47159.1 RLU27049.1 GDKW01000497 JAI56098.1 ACPB03017681 GAHY01000453 JAA77057.1 KZ288193 PBC33992.1 GEDC01024965 GEDC01013149 GEDC01004819 JAS12333.1 JAS24149.1 JAS32479.1 GFTR01006306 JAW10120.1 KK852969 KDR13109.1 GALA01001145 JAA93707.1 KQ435859 KOX70570.1 GL442548 EFN63308.1 GBHO01030053 GBHO01030052 GBRD01011961 GBRD01011960 GDHC01014561 GDHC01008306 JAG13551.1 JAG13552.1 JAG53863.1 JAQ04068.1 JAQ10323.1 GBGD01001456 JAC87433.1 KQ435007 KZC13531.1 GBBI01000791 JAC17921.1 KQ976818 KYN08113.1 KQ978782 KYN28391.1 KQ976490 KYM83468.1 GL888575 EGI59399.1 KQ982482 KYQ55946.1 KQ981424 KYN41821.1 ADTU01018078 CVRI01000015 CRK90032.1 GANO01000705 JAB59166.1 GAMD01002999 JAA98591.1 LJIG01016388 KRT80751.1 ADMH02002067 ETN59670.1 KQ971338 EFA01900.1 GGFJ01005008 MBW54149.1 GGFK01001742 MBW35063.1 GGFJ01005009 MBW54150.1 GFDF01004793 JAV09291.1 GGFM01000737 MBW21488.1 ATLV01020054 KE525293 KFB44785.1 APGK01008952 APGK01020978 APGK01051622 KB741197 KB740274 KB737822 KB632308 ENN72983.1 ENN80970.1 ENN82908.1 ERL91965.1 GGFM01000762 MBW21513.1 KQ414778 KOC61218.1 JXUM01044847 KQ561415 KXJ78656.1 UFQS01000027 UFQT01000027 SSW97719.1 SSX18105.1 GAPW01001401 JAC12197.1 JRES01001651 KNC21155.1 AXCM01008583 AAAB01008816 EAA05225.2 APCN01005436 CH477188 EAT48834.1 KA646479 AFP61108.1 GFXV01006290 MBW18095.1 GFDL01011125 JAV23920.1 AXCN02001714 DS231818 EDS41322.1 GEZM01063108 GEZM01063107 JAV69304.1 ABLF02031739 ABLF02031741 LBMM01000532 KMQ98121.1 DS235271 EEB14222.1 GAMC01016950 GAMC01016949 JAB89606.1 JXJN01023561 GAKP01017268 GAKP01017267 JAC41684.1 GBXI01004496 JAD09796.1 GBRD01017772 GBRD01011959 JAG53865.1 GDHF01031099 GDHF01028759 GDHF01028251 GDHF01022750 GDHF01022203 GDHF01017863 GDHF01015529 GDHF01012742 GDHF01008220 GDHF01006614 GDHF01005950 JAI21215.1 JAI23555.1 JAI24063.1 JAI29564.1 JAI30111.1 JAI34451.1 JAI36785.1 JAI39572.1 JAI44094.1 JAI45700.1 JAI46364.1 CH933811 EDW06419.1 CH964239 EDW82462.1 CH940655 EDW66380.1 AJWK01026604 AJWK01026605 AJWK01026606 OUUW01000011 SPP86446.1
Proteomes
UP000053268
UP000053240
UP000007151
UP000202317
UP000218220
UP000208100
+ More
UP000235965 UP000053097 UP000279307 UP000015103 UP000242457 UP000005203 UP000027135 UP000002358 UP000053105 UP000000311 UP000076502 UP000078542 UP000192223 UP000078492 UP000078540 UP000007755 UP000075809 UP000078541 UP000005205 UP000183832 UP000000673 UP000007266 UP000030765 UP000019118 UP000030742 UP000069272 UP000053825 UP000095300 UP000075885 UP000069940 UP000249989 UP000075880 UP000075884 UP000076408 UP000037069 UP000075902 UP000075900 UP000075920 UP000075883 UP000076407 UP000075903 UP000075882 UP000007062 UP000075840 UP000008820 UP000095301 UP000075881 UP000075886 UP000002320 UP000007819 UP000036403 UP000009046 UP000091820 UP000092443 UP000092460 UP000092445 UP000078200 UP000009192 UP000079169 UP000007798 UP000008792 UP000092461 UP000268350
UP000235965 UP000053097 UP000279307 UP000015103 UP000242457 UP000005203 UP000027135 UP000002358 UP000053105 UP000000311 UP000076502 UP000078542 UP000192223 UP000078492 UP000078540 UP000007755 UP000075809 UP000078541 UP000005205 UP000183832 UP000000673 UP000007266 UP000030765 UP000019118 UP000030742 UP000069272 UP000053825 UP000095300 UP000075885 UP000069940 UP000249989 UP000075880 UP000075884 UP000076408 UP000037069 UP000075902 UP000075900 UP000075920 UP000075883 UP000076407 UP000075903 UP000075882 UP000007062 UP000075840 UP000008820 UP000095301 UP000075881 UP000075886 UP000002320 UP000007819 UP000036403 UP000009046 UP000091820 UP000092443 UP000092460 UP000092445 UP000078200 UP000009192 UP000079169 UP000007798 UP000008792 UP000092461 UP000268350
PRIDE
Interpro
IPR003307
W2_domain
+ More
IPR016190 Transl_init_fac_IF2/IF5_Zn-bd
IPR016024 ARM-type_fold
IPR016189 Transl_init_fac_IF2/IF5_N
IPR016021 MIF4-like_sf
IPR002735 Transl_init_fac_IF2/IF5
IPR042089 Peptidase_M13_dom_2
IPR008753 Peptidase_M13_N
IPR024079 MetalloPept_cat_dom_sf
IPR018497 Peptidase_M13_C
IPR000718 Peptidase_M13
IPR016190 Transl_init_fac_IF2/IF5_Zn-bd
IPR016024 ARM-type_fold
IPR016189 Transl_init_fac_IF2/IF5_N
IPR016021 MIF4-like_sf
IPR002735 Transl_init_fac_IF2/IF5
IPR042089 Peptidase_M13_dom_2
IPR008753 Peptidase_M13_N
IPR024079 MetalloPept_cat_dom_sf
IPR018497 Peptidase_M13_C
IPR000718 Peptidase_M13
Gene 3D
CDD
ProteinModelPortal
Q0ZB74
A0A2H1VIU9
A0A1E1W4Z3
A0A194QME0
A0A194RIA0
A0A212EGP5
+ More
Q1A4T6 A0A2A4K9U5 S5N3Z2 S4PFZ8 A0A1B6MII3 A0A0K8TRP3 A0A0C9Q139 T1D408 A0A2J7PWA2 A0A026VWF8 A0A0P4VTX6 R4FNT1 A0A2A3ERU2 A0A088AP20 A0A1B6E3K8 A0A224XNV5 A0A067QT43 T1E2M2 K7J337 A0A0M8ZT33 E2ATE1 A0A0A9X1L2 A0A069DTN5 A0A154PNS0 A0A023F8Y9 A0A195D5F3 A0A1W4X3Q3 A0A195EJS1 A0A195BFU0 F4X2B7 A0A151X6D1 A0A195FN48 A0A158NJN0 A0A1J1HPP9 U5EXY4 T1DPV6 A0A0T6AZX0 W5J9Y9 D2A3J2 A0A2M4BM37 A0A2M4A2P3 A0A2M4BM36 A0A1L8DSB3 A0A2M3YZ48 A0A084W3J1 N6UGW8 A0A182FVI3 A0A2M3YZ22 A0A0L7QRG6 A0A1I8PQZ4 A0A182PI91 A0A182G5X2 A0A182J216 A0A336JZ43 A0A182MXI5 A0A182YEB8 A0A023ESH2 A0A0L0BM06 A0A182TDI3 A0A182RCZ2 A0A182WDE1 A0A182ME45 A0A1I8JW08 A0A182VL43 A0A182LHN7 Q7QHJ0 A0A182HFX9 Q17PW1 T1PCU9 A0A2H8TV07 A0A1Q3F8L5 A0A182KC71 A0A182QXD1 B0W0M0 A0A1Y1L6V3 J9K088 A0A0J7L661 E0VLG6 W8B8V3 A0A1A9WSZ9 A0A1A9X8Q0 A0A1B0C0E7 A0A034VGT6 A0A0A1XFT7 A0A1A9ZWY2 A0A0K8SM34 A0A0K8VE40 A0A1A9UQE2 B4L5G0 A0A3Q0JG87 B4NCA3 B4MAX1 A0A1B0CSU2 A0A3B0KKX5
Q1A4T6 A0A2A4K9U5 S5N3Z2 S4PFZ8 A0A1B6MII3 A0A0K8TRP3 A0A0C9Q139 T1D408 A0A2J7PWA2 A0A026VWF8 A0A0P4VTX6 R4FNT1 A0A2A3ERU2 A0A088AP20 A0A1B6E3K8 A0A224XNV5 A0A067QT43 T1E2M2 K7J337 A0A0M8ZT33 E2ATE1 A0A0A9X1L2 A0A069DTN5 A0A154PNS0 A0A023F8Y9 A0A195D5F3 A0A1W4X3Q3 A0A195EJS1 A0A195BFU0 F4X2B7 A0A151X6D1 A0A195FN48 A0A158NJN0 A0A1J1HPP9 U5EXY4 T1DPV6 A0A0T6AZX0 W5J9Y9 D2A3J2 A0A2M4BM37 A0A2M4A2P3 A0A2M4BM36 A0A1L8DSB3 A0A2M3YZ48 A0A084W3J1 N6UGW8 A0A182FVI3 A0A2M3YZ22 A0A0L7QRG6 A0A1I8PQZ4 A0A182PI91 A0A182G5X2 A0A182J216 A0A336JZ43 A0A182MXI5 A0A182YEB8 A0A023ESH2 A0A0L0BM06 A0A182TDI3 A0A182RCZ2 A0A182WDE1 A0A182ME45 A0A1I8JW08 A0A182VL43 A0A182LHN7 Q7QHJ0 A0A182HFX9 Q17PW1 T1PCU9 A0A2H8TV07 A0A1Q3F8L5 A0A182KC71 A0A182QXD1 B0W0M0 A0A1Y1L6V3 J9K088 A0A0J7L661 E0VLG6 W8B8V3 A0A1A9WSZ9 A0A1A9X8Q0 A0A1B0C0E7 A0A034VGT6 A0A0A1XFT7 A0A1A9ZWY2 A0A0K8SM34 A0A0K8VE40 A0A1A9UQE2 B4L5G0 A0A3Q0JG87 B4NCA3 B4MAX1 A0A1B0CSU2 A0A3B0KKX5
PDB
2E9H
E-value=6.0739e-65,
Score=629
Ontologies
KEGG
PATHWAY
GO
PANTHER
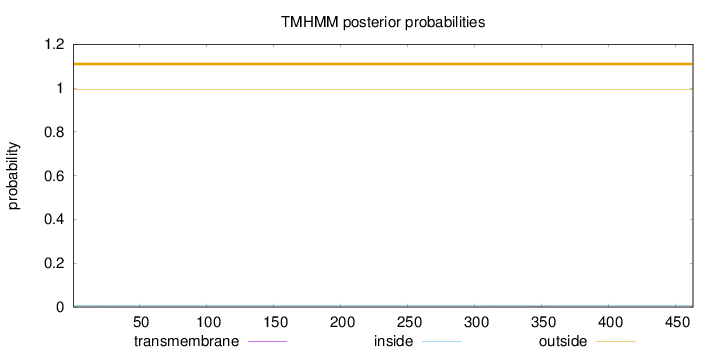
Topology
Length:
463
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00419
Exp number, first 60 AAs:
0.00043
Total prob of N-in:
0.00537
outside
1 - 463
Population Genetic Test Statistics
Pi
193.991015
Theta
169.328022
Tajima's D
-1.621794
CLR
0.589103
CSRT
0.0457977101144943
Interpretation
Possibly Positive selection
