Gene
KWMTBOMO11102
Pre Gene Modal
BGIBMGA008573
Annotation
PREDICTED:_poly(A)_RNA_polymerase?_mitochondrial_[Bombyx_mori]
Full name
Poly(A) RNA polymerase, mitochondrial
+ More
Aspartate aminotransferase
Aspartate aminotransferase
Location in the cell
Nuclear Reliability : 2.38
Sequence
CDS
ATGTCTGTGTTATTTCAAAGTATAATAAATACTGGAAAACCTTATTCATTAAGACAAATAATCAGCAAATCTACAAGAATCTGCAAATGTTTCTCAAAAAGAAAATTTTCAGATATTTCAAGGAAGTTTTTATCATTTGATGAAGTCGCCGCGCAGAGAAGGGCTGAAGCAAAACGAAGTCTAGTCGTTCAAGTCAATTCAGAAGCATCTTTTAATGAATTGTACGGATATTGTTCTAAATATGCTTCCGTCAATGGTATACATCATTACAAAAATTCTGGAGACGAGCATTTTATGCTCATCGAGTTCAAAACTGAAGAAGAAAGTAAGAATGTCATACACTCCTGTAGCTCACATCAAAAAGATGTTGACGTTATGGCTTTACATTCACCATTTCTATGGTTCAGAGCAGTCTCAGGAAAGAAAGAAGCGTTTCAGATTCCTAAATCACAACTCATGACGACAAAAGGTTGTAATTCAATCGATGAAGATAACTTAATTGAAGAGTTAGCAAATTGTGAGGATGTGTCAGAACAAATTCAACAGATGCACGATAGGACAGCTTTGAATGATCTAGGTGTGAGATTGAGGTATATGGTAGCAAGGCAGATAGAAAGGATATTTGAGAGCCTGTATGCCAATATAGTTGTGAGACCATTCGGGTCATCTGTGAATGGTTTTGGAAAGATGGGCTGTGACTTAGACTTGGTGCTTACCAATATCCTGAATGAAGAAATGTGTGACTCATCAAAGAGATTGGTATATCAAGAGAAGAAGTGCGAAGGTGGTCGCAGTCCTTGGCAGCGACACATGGAGCTAGTTGGAGAATTGTTAGAACTAAGAGTGCCTGGTGCTACTAGAGTACAGAGGATACTGCATGCAAGAGTCCCCATTGTAAAATATGCCCATGAATTCACTGATGTTGATTGCGATCTTTGTTATAATAATATGTCTGGAGTATACATGTCAGAATTGCTTTGGCTCTACGGTTCTCTGGACCCCAGGGTTCGACCGCTCACATTTGTAGTCCGTCGATGGGCCCAAACAGTGGGCCTCACCAACCAACACCCTGGGCGGTGGATCACAAACTTCCCACTCACTTTGATGGTACTGTTCTTTCTCCAACAGAAAGGCAAGGATGGATGTGTTCTACCATCATTGAAAACTCTTGTTCAATATGCTGGAAAAGAAGATATTAGATTAGCAGAAGAGAATCTAAACTGTACATTCCTGAGAGATATGAATAAATTGCCAACGGACACGTACGGACAAAATAAAGCTAATCTAGAAACGTTACTGCTACAGTTTTTCGAGTTTTACGCTCAATTCGACTTTCATGACAAAGCTATATCGATTAACGAGGGGGTGGCTATACGAAAGCCAAACACTCTACCATTGTACATAGTGAATCCTTTAGAACAAGCCTTGAATGTTAGCCGGAATGTGAGTTACGAGGAGTGCGAGAGGTTGAAACTAGAGGTCCGCAACGCAGCGTGGCTGTTAGATGCAGGTTTGGATGGTAAGAAAACTGACGGGTGGGGCATATTGGGCCTTATTGAGAGAAAAAGCACCAGGGGCCTAAAAAATCTGCTGAAAGTAGGAAATTCACATCGGCTTGTCTCCGTGAAGGATTTATTCAACAACGATGATGACAAAATTGAGAAAGTGAAGGCGAAAAGAGATAGTGAAATGAAACAGGTGCTCGCTCAAGTTGATAATAAACCAAAGGAGAAAATTAAGTTCAAGAACACGCAAGTTGCTAGTGAAGTTTATAGGATACGTAGGGATAAAATTATATGA
Protein
MSVLFQSIINTGKPYSLRQIISKSTRICKCFSKRKFSDISRKFLSFDEVAAQRRAEAKRSLVVQVNSEASFNELYGYCSKYASVNGIHHYKNSGDEHFMLIEFKTEEESKNVIHSCSSHQKDVDVMALHSPFLWFRAVSGKKEAFQIPKSQLMTTKGCNSIDEDNLIEELANCEDVSEQIQQMHDRTALNDLGVRLRYMVARQIERIFESLYANIVVRPFGSSVNGFGKMGCDLDLVLTNILNEEMCDSSKRLVYQEKKCEGGRSPWQRHMELVGELLELRVPGATRVQRILHARVPIVKYAHEFTDVDCDLCYNNMSGVYMSELLWLYGSLDPRVRPLTFVVRRWAQTVGLTNQHPGRWITNFPLTLMVLFFLQQKGKDGCVLPSLKTLVQYAGKEDIRLAEENLNCTFLRDMNKLPTDTYGQNKANLETLLLQFFEFYAQFDFHDKAISINEGVAIRKPNTLPLYIVNPLEQALNVSRNVSYEECERLKLEVRNAAWLLDAGLDGKKTDGWGILGLIERKSTRGLKNLLKVGNSHRLVSVKDLFNNDDDKIEKVKAKRDSEMKQVLAQVDNKPKEKIKFKNTQVASEVYRIRRDKII
Summary
Description
Polymerase that creates the 3' poly(A) tail of mitochondrial transcripts. This is not required for transcript stability or translation but may maintain mRNA integrity by protecting 3' termini from degradation.
Catalytic Activity
ATP + RNA(n) = diphosphate + RNA(n)-3'-adenine ribonucleotide
2-oxoglutarate + L-aspartate = L-glutamate + oxaloacetate
2-oxoglutarate + L-aspartate = L-glutamate + oxaloacetate
Subunit
Homodimer.
Miscellaneous
In eukaryotes there are cytoplasmic, mitochondrial and chloroplastic isozymes.
Similarity
Belongs to the DNA polymerase type-B-like family.
Keywords
ATP-binding
Complete proteome
Magnesium
Manganese
Metal-binding
Mitochondrion
mRNA processing
Nucleotide-binding
Nucleotidyltransferase
Reference proteome
RNA-binding
Transferase
Transit peptide
Feature
chain Poly(A) RNA polymerase, mitochondrial
Uniprot
H9JGC6
A0A2H1V5H0
A0A194RIM7
A0A194QGL1
A0A0L7LS96
U5ES50
+ More
B4GTN0 Q29IZ3 D6WRJ4 N6UHN1 U4UI64 A0A0K8U570 A0A3B0JW34 A0A1A9WCN3 A0A1W4VES3 B4PXN3 A0A034WU74 B4N1T4 W8C4A9 W8BIL5 A0A0A1X914 A0A0T6AVL1 B3MRE7 A0A1B0FFX7 A0A1A9VCS1 A0A1I8MY71 O46102 A0A1A9ZER3 T1PJZ1 B4L3W0 A0A0L0CP49 A0A1Y1L0P9 B4JLB0 A0A1A9XBM5 A0A1B0ATF2 B3P9G5 B4M3J7 A0A1I8PNX8 J9K7W6 A0A067QQB6 A0A2S2NMM5 B0W498 K7IRY0 A0A1S4EXH1 A0A336MLC6 A0A182VQL9 Q17MJ9 A0A336N5X3 A0A182MMM3 A0A2H8TGA2 A0A1S4GI68 A0A182X1Y7 A0A182RHY0 A0A182HJF5 A0A1B6MF47 A0A182TGJ3 A0A182UWT2 A0A1Q3F3W0 A0A182HDM3 Q7Q0J0 A0A336LRV6 A0A182YK94 A0A182JSN4 A0A182P5R6 A0A1J1HJX9 A0A182INP7 A0A1B0CAQ6 A0A182NL64 A0A1B0DAF9 W5J3W8 A0A2S2QAC1 A0A1S4EPJ9 A0A3Q0JG48 A0A182FQP7 A0A182QS77 A0A1B6EP16 A0A084VQL3 F4WXD2 A0A182LCR8 A0A158NKT9 E2C3E6 A0A151IS06 E9ICS1 A0A151X8B2 A0A1B6CKY0 A0A0V0GB48 A0A023ETY7 E2ALG8 A0A0A9ZGE8 A0A023F1M2 A0A195CYI5 A0A195F541 A0A3L8DT43 A0A026VZ68
B4GTN0 Q29IZ3 D6WRJ4 N6UHN1 U4UI64 A0A0K8U570 A0A3B0JW34 A0A1A9WCN3 A0A1W4VES3 B4PXN3 A0A034WU74 B4N1T4 W8C4A9 W8BIL5 A0A0A1X914 A0A0T6AVL1 B3MRE7 A0A1B0FFX7 A0A1A9VCS1 A0A1I8MY71 O46102 A0A1A9ZER3 T1PJZ1 B4L3W0 A0A0L0CP49 A0A1Y1L0P9 B4JLB0 A0A1A9XBM5 A0A1B0ATF2 B3P9G5 B4M3J7 A0A1I8PNX8 J9K7W6 A0A067QQB6 A0A2S2NMM5 B0W498 K7IRY0 A0A1S4EXH1 A0A336MLC6 A0A182VQL9 Q17MJ9 A0A336N5X3 A0A182MMM3 A0A2H8TGA2 A0A1S4GI68 A0A182X1Y7 A0A182RHY0 A0A182HJF5 A0A1B6MF47 A0A182TGJ3 A0A182UWT2 A0A1Q3F3W0 A0A182HDM3 Q7Q0J0 A0A336LRV6 A0A182YK94 A0A182JSN4 A0A182P5R6 A0A1J1HJX9 A0A182INP7 A0A1B0CAQ6 A0A182NL64 A0A1B0DAF9 W5J3W8 A0A2S2QAC1 A0A1S4EPJ9 A0A3Q0JG48 A0A182FQP7 A0A182QS77 A0A1B6EP16 A0A084VQL3 F4WXD2 A0A182LCR8 A0A158NKT9 E2C3E6 A0A151IS06 E9ICS1 A0A151X8B2 A0A1B6CKY0 A0A0V0GB48 A0A023ETY7 E2ALG8 A0A0A9ZGE8 A0A023F1M2 A0A195CYI5 A0A195F541 A0A3L8DT43 A0A026VZ68
EC Number
2.7.7.19
2.6.1.1
2.6.1.1
Pubmed
19121390
26354079
26227816
17994087
15632085
18362917
+ More
19820115 23537049 17550304 25348373 24495485 25830018 25315136 10731137 10731132 12537572 12537569 27176048 26108605 28004739 24845553 20075255 17510324 12364791 26483478 25244985 20920257 23761445 24438588 21719571 20966253 21347285 20798317 21282665 24945155 25401762 25474469 30249741 24508170
19820115 23537049 17550304 25348373 24495485 25830018 25315136 10731137 10731132 12537572 12537569 27176048 26108605 28004739 24845553 20075255 17510324 12364791 26483478 25244985 20920257 23761445 24438588 21719571 20966253 21347285 20798317 21282665 24945155 25401762 25474469 30249741 24508170
EMBL
BABH01002463
ODYU01000763
SOQ36039.1
KQ460154
KPJ17294.1
KQ458880
+ More
KPJ04642.1 JTDY01000200 KOB78322.1 GANO01003406 JAB56465.1 CH479190 EDW25900.1 CH379063 EAL32510.3 KQ971351 EFA06440.1 APGK01019954 KB740150 ENN81250.1 KB632173 ERL89575.1 GDHF01030603 JAI21711.1 OUUW01000011 SPP86264.1 CM000162 EDX00886.1 GAKP01001232 JAC57720.1 CH963925 EDW78323.2 GAMC01009691 JAB96864.1 GAMC01009692 JAB96863.1 GBXI01006513 JAD07779.1 LJIG01022704 KRT79145.1 CH902622 EDV34352.2 CCAG010008264 AL022018 AE014298 AY119282 BT132966 AAM51142.1 AEW48257.1 CAA17688.2 KA648979 AFP63608.1 CH933810 EDW07238.1 JRES01000104 KNC34110.1 GEZM01068036 JAV67249.1 CH916370 EDW00363.1 JXJN01003292 CH954183 EDV45461.1 CH940651 EDW65372.1 ABLF02034857 KK853582 KDR06507.1 GGMR01005583 MBY18202.1 DS231836 EDS33183.1 UFQT01001585 SSX31066.1 CH477205 EAT47925.1 UFQT01002692 SSX33948.1 AXCM01001179 GFXV01001274 MBW13079.1 AAAB01008794 AAAB01008982 APCN01002160 GEBQ01005462 JAT34515.1 GFDL01012801 JAV22244.1 JXUM01129974 KQ567517 KXJ69395.1 EAA14654.4 UFQS01004354 UFQT01004354 SSX16342.1 SSX35661.1 CVRI01000004 CRK87716.1 AJWK01004158 AJVK01013292 ADMH02002181 ETN58018.1 GGMS01005277 MBY74480.1 AXCN02000083 GECZ01030166 JAS39603.1 ATLV01015250 KE525004 KFB40257.1 GL888424 EGI61098.1 ADTU01002130 GL452320 EFN77484.1 KQ981102 KYN09461.1 GL762354 EFZ21622.1 KQ982431 KYQ56550.1 GEDC01023260 JAS14038.1 GECL01000801 JAP05323.1 GAPW01001237 JAC12361.1 GL440607 EFN65726.1 GBHO01000468 JAG43136.1 GBBI01003818 JAC14894.1 KQ977110 KYN05718.1 KQ981805 KYN35578.1 QOIP01000005 RLU22938.1 KK107570 EZA48761.1
KPJ04642.1 JTDY01000200 KOB78322.1 GANO01003406 JAB56465.1 CH479190 EDW25900.1 CH379063 EAL32510.3 KQ971351 EFA06440.1 APGK01019954 KB740150 ENN81250.1 KB632173 ERL89575.1 GDHF01030603 JAI21711.1 OUUW01000011 SPP86264.1 CM000162 EDX00886.1 GAKP01001232 JAC57720.1 CH963925 EDW78323.2 GAMC01009691 JAB96864.1 GAMC01009692 JAB96863.1 GBXI01006513 JAD07779.1 LJIG01022704 KRT79145.1 CH902622 EDV34352.2 CCAG010008264 AL022018 AE014298 AY119282 BT132966 AAM51142.1 AEW48257.1 CAA17688.2 KA648979 AFP63608.1 CH933810 EDW07238.1 JRES01000104 KNC34110.1 GEZM01068036 JAV67249.1 CH916370 EDW00363.1 JXJN01003292 CH954183 EDV45461.1 CH940651 EDW65372.1 ABLF02034857 KK853582 KDR06507.1 GGMR01005583 MBY18202.1 DS231836 EDS33183.1 UFQT01001585 SSX31066.1 CH477205 EAT47925.1 UFQT01002692 SSX33948.1 AXCM01001179 GFXV01001274 MBW13079.1 AAAB01008794 AAAB01008982 APCN01002160 GEBQ01005462 JAT34515.1 GFDL01012801 JAV22244.1 JXUM01129974 KQ567517 KXJ69395.1 EAA14654.4 UFQS01004354 UFQT01004354 SSX16342.1 SSX35661.1 CVRI01000004 CRK87716.1 AJWK01004158 AJVK01013292 ADMH02002181 ETN58018.1 GGMS01005277 MBY74480.1 AXCN02000083 GECZ01030166 JAS39603.1 ATLV01015250 KE525004 KFB40257.1 GL888424 EGI61098.1 ADTU01002130 GL452320 EFN77484.1 KQ981102 KYN09461.1 GL762354 EFZ21622.1 KQ982431 KYQ56550.1 GEDC01023260 JAS14038.1 GECL01000801 JAP05323.1 GAPW01001237 JAC12361.1 GL440607 EFN65726.1 GBHO01000468 JAG43136.1 GBBI01003818 JAC14894.1 KQ977110 KYN05718.1 KQ981805 KYN35578.1 QOIP01000005 RLU22938.1 KK107570 EZA48761.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000037510
UP000008744
UP000001819
+ More
UP000007266 UP000019118 UP000030742 UP000268350 UP000091820 UP000192221 UP000002282 UP000007798 UP000007801 UP000092444 UP000078200 UP000095301 UP000000803 UP000092445 UP000009192 UP000037069 UP000001070 UP000092443 UP000092460 UP000008711 UP000008792 UP000095300 UP000007819 UP000027135 UP000002320 UP000002358 UP000075920 UP000008820 UP000075883 UP000076407 UP000075900 UP000075840 UP000075902 UP000075903 UP000069940 UP000249989 UP000007062 UP000076408 UP000075881 UP000075885 UP000183832 UP000075880 UP000092461 UP000075884 UP000092462 UP000000673 UP000079169 UP000069272 UP000075886 UP000030765 UP000007755 UP000075882 UP000005205 UP000008237 UP000078492 UP000075809 UP000000311 UP000078542 UP000078541 UP000279307 UP000053097
UP000007266 UP000019118 UP000030742 UP000268350 UP000091820 UP000192221 UP000002282 UP000007798 UP000007801 UP000092444 UP000078200 UP000095301 UP000000803 UP000092445 UP000009192 UP000037069 UP000001070 UP000092443 UP000092460 UP000008711 UP000008792 UP000095300 UP000007819 UP000027135 UP000002320 UP000002358 UP000075920 UP000008820 UP000075883 UP000076407 UP000075900 UP000075840 UP000075902 UP000075903 UP000069940 UP000249989 UP000007062 UP000076408 UP000075881 UP000075885 UP000183832 UP000075880 UP000092461 UP000075884 UP000092462 UP000000673 UP000079169 UP000069272 UP000075886 UP000030765 UP000007755 UP000075882 UP000005205 UP000008237 UP000078492 UP000075809 UP000000311 UP000078542 UP000078541 UP000279307 UP000053097
Pfam
Interpro
IPR002058
PAP_assoc
+ More
IPR041252 RL
IPR035979 RBD_domain_sf
IPR002934 Polymerase_NTP_transf_dom
IPR015421 PyrdxlP-dep_Trfase_major
IPR015311 DFF40_C
IPR004839 Aminotransferase_I/II
IPR015424 PyrdxlP-dep_Trfase
IPR000683 Oxidoreductase_N
IPR000796 Asp_trans
IPR036291 NAD(P)-bd_dom_sf
IPR003508 CIDE-N_dom
IPR004838 NHTrfase_class1_PyrdxlP-BS
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR041252 RL
IPR035979 RBD_domain_sf
IPR002934 Polymerase_NTP_transf_dom
IPR015421 PyrdxlP-dep_Trfase_major
IPR015311 DFF40_C
IPR004839 Aminotransferase_I/II
IPR015424 PyrdxlP-dep_Trfase
IPR000683 Oxidoreductase_N
IPR000796 Asp_trans
IPR036291 NAD(P)-bd_dom_sf
IPR003508 CIDE-N_dom
IPR004838 NHTrfase_class1_PyrdxlP-BS
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR012677 Nucleotide-bd_a/b_plait_sf
Gene 3D
ProteinModelPortal
H9JGC6
A0A2H1V5H0
A0A194RIM7
A0A194QGL1
A0A0L7LS96
U5ES50
+ More
B4GTN0 Q29IZ3 D6WRJ4 N6UHN1 U4UI64 A0A0K8U570 A0A3B0JW34 A0A1A9WCN3 A0A1W4VES3 B4PXN3 A0A034WU74 B4N1T4 W8C4A9 W8BIL5 A0A0A1X914 A0A0T6AVL1 B3MRE7 A0A1B0FFX7 A0A1A9VCS1 A0A1I8MY71 O46102 A0A1A9ZER3 T1PJZ1 B4L3W0 A0A0L0CP49 A0A1Y1L0P9 B4JLB0 A0A1A9XBM5 A0A1B0ATF2 B3P9G5 B4M3J7 A0A1I8PNX8 J9K7W6 A0A067QQB6 A0A2S2NMM5 B0W498 K7IRY0 A0A1S4EXH1 A0A336MLC6 A0A182VQL9 Q17MJ9 A0A336N5X3 A0A182MMM3 A0A2H8TGA2 A0A1S4GI68 A0A182X1Y7 A0A182RHY0 A0A182HJF5 A0A1B6MF47 A0A182TGJ3 A0A182UWT2 A0A1Q3F3W0 A0A182HDM3 Q7Q0J0 A0A336LRV6 A0A182YK94 A0A182JSN4 A0A182P5R6 A0A1J1HJX9 A0A182INP7 A0A1B0CAQ6 A0A182NL64 A0A1B0DAF9 W5J3W8 A0A2S2QAC1 A0A1S4EPJ9 A0A3Q0JG48 A0A182FQP7 A0A182QS77 A0A1B6EP16 A0A084VQL3 F4WXD2 A0A182LCR8 A0A158NKT9 E2C3E6 A0A151IS06 E9ICS1 A0A151X8B2 A0A1B6CKY0 A0A0V0GB48 A0A023ETY7 E2ALG8 A0A0A9ZGE8 A0A023F1M2 A0A195CYI5 A0A195F541 A0A3L8DT43 A0A026VZ68
B4GTN0 Q29IZ3 D6WRJ4 N6UHN1 U4UI64 A0A0K8U570 A0A3B0JW34 A0A1A9WCN3 A0A1W4VES3 B4PXN3 A0A034WU74 B4N1T4 W8C4A9 W8BIL5 A0A0A1X914 A0A0T6AVL1 B3MRE7 A0A1B0FFX7 A0A1A9VCS1 A0A1I8MY71 O46102 A0A1A9ZER3 T1PJZ1 B4L3W0 A0A0L0CP49 A0A1Y1L0P9 B4JLB0 A0A1A9XBM5 A0A1B0ATF2 B3P9G5 B4M3J7 A0A1I8PNX8 J9K7W6 A0A067QQB6 A0A2S2NMM5 B0W498 K7IRY0 A0A1S4EXH1 A0A336MLC6 A0A182VQL9 Q17MJ9 A0A336N5X3 A0A182MMM3 A0A2H8TGA2 A0A1S4GI68 A0A182X1Y7 A0A182RHY0 A0A182HJF5 A0A1B6MF47 A0A182TGJ3 A0A182UWT2 A0A1Q3F3W0 A0A182HDM3 Q7Q0J0 A0A336LRV6 A0A182YK94 A0A182JSN4 A0A182P5R6 A0A1J1HJX9 A0A182INP7 A0A1B0CAQ6 A0A182NL64 A0A1B0DAF9 W5J3W8 A0A2S2QAC1 A0A1S4EPJ9 A0A3Q0JG48 A0A182FQP7 A0A182QS77 A0A1B6EP16 A0A084VQL3 F4WXD2 A0A182LCR8 A0A158NKT9 E2C3E6 A0A151IS06 E9ICS1 A0A151X8B2 A0A1B6CKY0 A0A0V0GB48 A0A023ETY7 E2ALG8 A0A0A9ZGE8 A0A023F1M2 A0A195CYI5 A0A195F541 A0A3L8DT43 A0A026VZ68
PDB
5A2Z
E-value=1.34017e-69,
Score=670
Ontologies
GO
PANTHER
Topology
Subcellular location
Mitochondrion
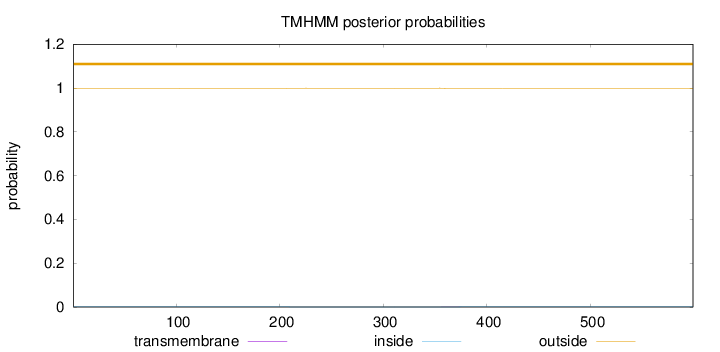
Length:
599
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01676
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00062
outside
1 - 599
Population Genetic Test Statistics
Pi
24.438812
Theta
155.976042
Tajima's D
-0.818608
CLR
265.726318
CSRT
0.170091495425229
Interpretation
Uncertain
