Pre Gene Modal
BGIBMGA008291
Annotation
Mitochondrial_ribosome_recycling_factor_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 1.153 Mitochondrial Reliability : 1.294 Nuclear Reliability : 1.654
Sequence
CDS
ATGGCAGTTCTCTCCGAACTTATACCAGTAGACAAGATGAAAGACCGCTGCAGTGCCGCTATTGAAAACATGAAGGCGGATTTTGCTAAACATTTGTCTATAAGATCAACCACTGGCTCGATAGACACAATACCAGTAAAATTCGAGGGCAAAGAATATGAACTACAAGAACTAGCTCAGATTGTAAGAAAGAATCCTAAAACGATAGTAATAAACTTCTCTTCTTTTCCCCAAGTCATACCGGAGGCCTTGAAAGCCATCAGTAGTTCTGGTCTAAATTTAAATCCACAACAAGATGGAACTACACTGTTTGTGCCTGTGCCTAAAGTCACAAAGGAGCACAGGGAAGCATTGGCTAAGAATGCTAAGGTGTTGTACATAAAGTGTAGGGATGCACTCAAAGACATACAAAATGAAAACATAAAGAAAATGAAGAAACAGACTGGTGTATCTGAAGATTTAATATTTAACGGAACGAAACAAATAAATGCCTTGTGTGAAAAATATCTATTAGAAGCGAGAAGTATTTATGATGCCAAATGTGTTGAATTAGTTGGGAAGTAG
Protein
MAVLSELIPVDKMKDRCSAAIENMKADFAKHLSIRSTTGSIDTIPVKFEGKEYELQELAQIVRKNPKTIVINFSSFPQVIPEALKAISSSGLNLNPQQDGTTLFVPVPKVTKEHREALAKNAKVLYIKCRDALKDIQNENIKKMKKQTGVSEDLIFNGTKQINALCEKYLLEARSIYDAKCVELVGK
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Uniprot
H9JFJ4
A0A0L7KP94
A0A194QGH9
A0A194RIY1
A0A2A4K0P3
A0A212FKK7
+ More
A0A2H1V5B3 A0A067RE93 A0A182WI37 A0A2J7RCB9 Q16WG4 A0A1S4FLR2 D6WBQ5 A0A182MBM7 A0A182N7G1 A0A182GF64 A0A2P8ZND4 A0A182IWI8 A0A182K0Q2 A0A182TN27 A0A0A1XLF1 A0A182I6D4 Q7QIE2 A0A182V6G6 A0A182L400 A0A182R6P6 A0A0K8V3I3 U5EVR6 A0A1Q3F6V6 A0A034VNS6 A0A1I8P6V6 A0A182X274 B4KX20 A0A0C9QDC5 A0A1I8NBK3 B0WC75 A0A0M4EN57 A0A182SD24 J3JY02 W8CB20 B4LE26 B4IYH5 A0A2M4BZ13 A0A2M4CU16 A0A2M4CU53 A0A2M4BZG8 A0A0K8TL14 T1PIC7 A0A2M4BXQ1 N6T5R9 Q29F88 Q9VSW4 A0A0J9RS36 A0A1A9W4C6 B4PEZ0 B3NCE2 B4MN38 A0A182FFJ5 A0A1W4VP28 B3M6E3 K7J7S1 R4UWU4 A0A1B6IJ35 Q8T0M6 A0A2M3ZJD9 A0A0L0CQD6 A0A1I8P6U0 A0A1A9YIA2 A0A1B6EVL2 A0A0T6BBX2 A0A1B0AVT3 A0A232F347 A0A1A9UGD0 A0A1B6KM94 A0A1W4XSC9 A0A1B0G3B0 A0A1L8DRK8 A0A1I8P717 X1XBB2 A0A1J1IGT5 A0A1Y1NAN7 A0A2S2Q0Y1 J9JR21 A0A1B0GPP0 E2AZY8 A0A2H8TKM8 A0A336MG77 A0A336MRP8 A0A1B6E5S0 A0A1B6EC03 A0A224XWZ4 A0A1S4EL10 A0A146LYX7 E2CA31 A0A151WTR9 E9IKN9 A0A3R7PAG5 A0A1B0CQE4 A0A195D3C3 A0A0P5TU04
A0A2H1V5B3 A0A067RE93 A0A182WI37 A0A2J7RCB9 Q16WG4 A0A1S4FLR2 D6WBQ5 A0A182MBM7 A0A182N7G1 A0A182GF64 A0A2P8ZND4 A0A182IWI8 A0A182K0Q2 A0A182TN27 A0A0A1XLF1 A0A182I6D4 Q7QIE2 A0A182V6G6 A0A182L400 A0A182R6P6 A0A0K8V3I3 U5EVR6 A0A1Q3F6V6 A0A034VNS6 A0A1I8P6V6 A0A182X274 B4KX20 A0A0C9QDC5 A0A1I8NBK3 B0WC75 A0A0M4EN57 A0A182SD24 J3JY02 W8CB20 B4LE26 B4IYH5 A0A2M4BZ13 A0A2M4CU16 A0A2M4CU53 A0A2M4BZG8 A0A0K8TL14 T1PIC7 A0A2M4BXQ1 N6T5R9 Q29F88 Q9VSW4 A0A0J9RS36 A0A1A9W4C6 B4PEZ0 B3NCE2 B4MN38 A0A182FFJ5 A0A1W4VP28 B3M6E3 K7J7S1 R4UWU4 A0A1B6IJ35 Q8T0M6 A0A2M3ZJD9 A0A0L0CQD6 A0A1I8P6U0 A0A1A9YIA2 A0A1B6EVL2 A0A0T6BBX2 A0A1B0AVT3 A0A232F347 A0A1A9UGD0 A0A1B6KM94 A0A1W4XSC9 A0A1B0G3B0 A0A1L8DRK8 A0A1I8P717 X1XBB2 A0A1J1IGT5 A0A1Y1NAN7 A0A2S2Q0Y1 J9JR21 A0A1B0GPP0 E2AZY8 A0A2H8TKM8 A0A336MG77 A0A336MRP8 A0A1B6E5S0 A0A1B6EC03 A0A224XWZ4 A0A1S4EL10 A0A146LYX7 E2CA31 A0A151WTR9 E9IKN9 A0A3R7PAG5 A0A1B0CQE4 A0A195D3C3 A0A0P5TU04
Pubmed
19121390
26227816
26354079
22118469
24845553
17510324
+ More
18362917 19820115 26483478 29403074 25830018 12364791 14747013 17210077 20966253 25348373 17994087 25315136 22516182 24495485 26369729 23537049 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17550304 20075255 26108605 28648823 28004739 20798317 26823975 21282665
18362917 19820115 26483478 29403074 25830018 12364791 14747013 17210077 20966253 25348373 17994087 25315136 22516182 24495485 26369729 23537049 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17550304 20075255 26108605 28648823 28004739 20798317 26823975 21282665
EMBL
BABH01002459
JTDY01008045
KOB64779.1
KQ458880
KPJ04643.1
KQ460154
+ More
KPJ17295.1 NWSH01000306 PCG77586.1 AGBW02008030 OWR54266.1 ODYU01000763 SOQ36040.1 KK852522 KDR22082.1 NEVH01005885 PNF38468.1 CH477566 EAT38932.1 KQ971313 EEZ98734.1 AXCM01002040 JXUM01059293 KQ562045 KXJ76822.1 PYGN01000010 PSN58022.1 GBXI01002904 JAD11388.1 APCN01003620 AAAB01008807 EAA04181.2 GDHF01018908 JAI33406.1 GANO01001748 JAB58123.1 GFDL01011768 JAV23277.1 GAKP01015769 GAKP01015762 JAC43183.1 CH933809 EDW19663.1 GBYB01001284 JAG71051.1 DS231886 EDS43320.1 CP012525 ALC43292.1 BT128125 AEE63086.1 GAMC01005221 JAC01335.1 CH940647 EDW70069.1 CH916366 EDV97648.1 GGFJ01009186 MBW58327.1 GGFL01004150 MBW68328.1 GGFL01004672 MBW68850.1 GGFJ01009334 MBW58475.1 GDAI01002536 JAI15067.1 KA648552 AFP63181.1 GGFJ01008714 MBW57855.1 APGK01042984 KB741011 ENN75539.1 CH379067 EAL31367.3 AE014296 BT044295 AAF50297.1 ACH92360.1 CM002912 KMY98603.1 CM000159 EDW93045.1 CH954178 EDV51172.1 CH963847 EDW73594.1 CH902618 EDV40792.1 AAZX01003361 KC740957 AGM32781.1 GECU01020793 JAS86913.1 AY069172 AAL39317.1 GGFM01007819 MBW28570.1 JRES01000080 KNC34377.1 GECZ01027754 JAS42015.1 LJIG01002131 KRT84809.1 JXJN01004388 NNAY01001113 OXU25095.1 GEBQ01027414 GEBQ01021880 GEBQ01013549 GEBQ01002416 JAT12563.1 JAT18097.1 JAT26428.1 JAT37561.1 CCAG010013743 GFDF01005094 JAV08990.1 ABLF02039150 CVRI01000050 CRK99416.1 GEZM01007730 JAV94971.1 GGMS01002224 MBY71427.1 ABLF02014514 AJVK01059901 GL444289 EFN60991.1 GFXV01002889 MBW14694.1 UFQS01001185 UFQT01001185 SSX09588.1 SSX29384.1 UFQS01002322 UFQT01002322 SSX13793.1 SSX33214.1 GEDC01004017 JAS33281.1 GEDC01001878 JAS35420.1 GFTR01003743 JAW12683.1 GDHC01006793 JAQ11836.1 GL453920 EFN75186.1 KQ982753 KYQ51234.1 GL764022 EFZ18864.1 QCYY01003116 ROT65233.1 AJWK01023464 AJWK01023465 KQ976885 KYN07397.1 GDIP01126904 LRGB01002851 JAL76810.1 KZS05975.1
KPJ17295.1 NWSH01000306 PCG77586.1 AGBW02008030 OWR54266.1 ODYU01000763 SOQ36040.1 KK852522 KDR22082.1 NEVH01005885 PNF38468.1 CH477566 EAT38932.1 KQ971313 EEZ98734.1 AXCM01002040 JXUM01059293 KQ562045 KXJ76822.1 PYGN01000010 PSN58022.1 GBXI01002904 JAD11388.1 APCN01003620 AAAB01008807 EAA04181.2 GDHF01018908 JAI33406.1 GANO01001748 JAB58123.1 GFDL01011768 JAV23277.1 GAKP01015769 GAKP01015762 JAC43183.1 CH933809 EDW19663.1 GBYB01001284 JAG71051.1 DS231886 EDS43320.1 CP012525 ALC43292.1 BT128125 AEE63086.1 GAMC01005221 JAC01335.1 CH940647 EDW70069.1 CH916366 EDV97648.1 GGFJ01009186 MBW58327.1 GGFL01004150 MBW68328.1 GGFL01004672 MBW68850.1 GGFJ01009334 MBW58475.1 GDAI01002536 JAI15067.1 KA648552 AFP63181.1 GGFJ01008714 MBW57855.1 APGK01042984 KB741011 ENN75539.1 CH379067 EAL31367.3 AE014296 BT044295 AAF50297.1 ACH92360.1 CM002912 KMY98603.1 CM000159 EDW93045.1 CH954178 EDV51172.1 CH963847 EDW73594.1 CH902618 EDV40792.1 AAZX01003361 KC740957 AGM32781.1 GECU01020793 JAS86913.1 AY069172 AAL39317.1 GGFM01007819 MBW28570.1 JRES01000080 KNC34377.1 GECZ01027754 JAS42015.1 LJIG01002131 KRT84809.1 JXJN01004388 NNAY01001113 OXU25095.1 GEBQ01027414 GEBQ01021880 GEBQ01013549 GEBQ01002416 JAT12563.1 JAT18097.1 JAT26428.1 JAT37561.1 CCAG010013743 GFDF01005094 JAV08990.1 ABLF02039150 CVRI01000050 CRK99416.1 GEZM01007730 JAV94971.1 GGMS01002224 MBY71427.1 ABLF02014514 AJVK01059901 GL444289 EFN60991.1 GFXV01002889 MBW14694.1 UFQS01001185 UFQT01001185 SSX09588.1 SSX29384.1 UFQS01002322 UFQT01002322 SSX13793.1 SSX33214.1 GEDC01004017 JAS33281.1 GEDC01001878 JAS35420.1 GFTR01003743 JAW12683.1 GDHC01006793 JAQ11836.1 GL453920 EFN75186.1 KQ982753 KYQ51234.1 GL764022 EFZ18864.1 QCYY01003116 ROT65233.1 AJWK01023464 AJWK01023465 KQ976885 KYN07397.1 GDIP01126904 LRGB01002851 JAL76810.1 KZS05975.1
Proteomes
UP000005204
UP000037510
UP000053268
UP000053240
UP000218220
UP000007151
+ More
UP000027135 UP000075920 UP000235965 UP000008820 UP000007266 UP000075883 UP000075884 UP000069940 UP000249989 UP000245037 UP000075880 UP000075881 UP000075902 UP000075840 UP000007062 UP000075903 UP000075882 UP000075900 UP000095300 UP000076407 UP000009192 UP000095301 UP000002320 UP000092553 UP000075901 UP000008792 UP000001070 UP000019118 UP000001819 UP000000803 UP000091820 UP000002282 UP000008711 UP000007798 UP000069272 UP000192221 UP000007801 UP000002358 UP000037069 UP000092443 UP000092460 UP000215335 UP000078200 UP000192223 UP000092444 UP000007819 UP000183832 UP000092462 UP000000311 UP000079169 UP000008237 UP000075809 UP000283509 UP000092461 UP000078542 UP000076858
UP000027135 UP000075920 UP000235965 UP000008820 UP000007266 UP000075883 UP000075884 UP000069940 UP000249989 UP000245037 UP000075880 UP000075881 UP000075902 UP000075840 UP000007062 UP000075903 UP000075882 UP000075900 UP000095300 UP000076407 UP000009192 UP000095301 UP000002320 UP000092553 UP000075901 UP000008792 UP000001070 UP000019118 UP000001819 UP000000803 UP000091820 UP000002282 UP000008711 UP000007798 UP000069272 UP000192221 UP000007801 UP000002358 UP000037069 UP000092443 UP000092460 UP000215335 UP000078200 UP000192223 UP000092444 UP000007819 UP000183832 UP000092462 UP000000311 UP000079169 UP000008237 UP000075809 UP000283509 UP000092461 UP000078542 UP000076858
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JFJ4
A0A0L7KP94
A0A194QGH9
A0A194RIY1
A0A2A4K0P3
A0A212FKK7
+ More
A0A2H1V5B3 A0A067RE93 A0A182WI37 A0A2J7RCB9 Q16WG4 A0A1S4FLR2 D6WBQ5 A0A182MBM7 A0A182N7G1 A0A182GF64 A0A2P8ZND4 A0A182IWI8 A0A182K0Q2 A0A182TN27 A0A0A1XLF1 A0A182I6D4 Q7QIE2 A0A182V6G6 A0A182L400 A0A182R6P6 A0A0K8V3I3 U5EVR6 A0A1Q3F6V6 A0A034VNS6 A0A1I8P6V6 A0A182X274 B4KX20 A0A0C9QDC5 A0A1I8NBK3 B0WC75 A0A0M4EN57 A0A182SD24 J3JY02 W8CB20 B4LE26 B4IYH5 A0A2M4BZ13 A0A2M4CU16 A0A2M4CU53 A0A2M4BZG8 A0A0K8TL14 T1PIC7 A0A2M4BXQ1 N6T5R9 Q29F88 Q9VSW4 A0A0J9RS36 A0A1A9W4C6 B4PEZ0 B3NCE2 B4MN38 A0A182FFJ5 A0A1W4VP28 B3M6E3 K7J7S1 R4UWU4 A0A1B6IJ35 Q8T0M6 A0A2M3ZJD9 A0A0L0CQD6 A0A1I8P6U0 A0A1A9YIA2 A0A1B6EVL2 A0A0T6BBX2 A0A1B0AVT3 A0A232F347 A0A1A9UGD0 A0A1B6KM94 A0A1W4XSC9 A0A1B0G3B0 A0A1L8DRK8 A0A1I8P717 X1XBB2 A0A1J1IGT5 A0A1Y1NAN7 A0A2S2Q0Y1 J9JR21 A0A1B0GPP0 E2AZY8 A0A2H8TKM8 A0A336MG77 A0A336MRP8 A0A1B6E5S0 A0A1B6EC03 A0A224XWZ4 A0A1S4EL10 A0A146LYX7 E2CA31 A0A151WTR9 E9IKN9 A0A3R7PAG5 A0A1B0CQE4 A0A195D3C3 A0A0P5TU04
A0A2H1V5B3 A0A067RE93 A0A182WI37 A0A2J7RCB9 Q16WG4 A0A1S4FLR2 D6WBQ5 A0A182MBM7 A0A182N7G1 A0A182GF64 A0A2P8ZND4 A0A182IWI8 A0A182K0Q2 A0A182TN27 A0A0A1XLF1 A0A182I6D4 Q7QIE2 A0A182V6G6 A0A182L400 A0A182R6P6 A0A0K8V3I3 U5EVR6 A0A1Q3F6V6 A0A034VNS6 A0A1I8P6V6 A0A182X274 B4KX20 A0A0C9QDC5 A0A1I8NBK3 B0WC75 A0A0M4EN57 A0A182SD24 J3JY02 W8CB20 B4LE26 B4IYH5 A0A2M4BZ13 A0A2M4CU16 A0A2M4CU53 A0A2M4BZG8 A0A0K8TL14 T1PIC7 A0A2M4BXQ1 N6T5R9 Q29F88 Q9VSW4 A0A0J9RS36 A0A1A9W4C6 B4PEZ0 B3NCE2 B4MN38 A0A182FFJ5 A0A1W4VP28 B3M6E3 K7J7S1 R4UWU4 A0A1B6IJ35 Q8T0M6 A0A2M3ZJD9 A0A0L0CQD6 A0A1I8P6U0 A0A1A9YIA2 A0A1B6EVL2 A0A0T6BBX2 A0A1B0AVT3 A0A232F347 A0A1A9UGD0 A0A1B6KM94 A0A1W4XSC9 A0A1B0G3B0 A0A1L8DRK8 A0A1I8P717 X1XBB2 A0A1J1IGT5 A0A1Y1NAN7 A0A2S2Q0Y1 J9JR21 A0A1B0GPP0 E2AZY8 A0A2H8TKM8 A0A336MG77 A0A336MRP8 A0A1B6E5S0 A0A1B6EC03 A0A224XWZ4 A0A1S4EL10 A0A146LYX7 E2CA31 A0A151WTR9 E9IKN9 A0A3R7PAG5 A0A1B0CQE4 A0A195D3C3 A0A0P5TU04
PDB
6NU2
E-value=2.13585e-25,
Score=283
Ontologies
GO
Topology
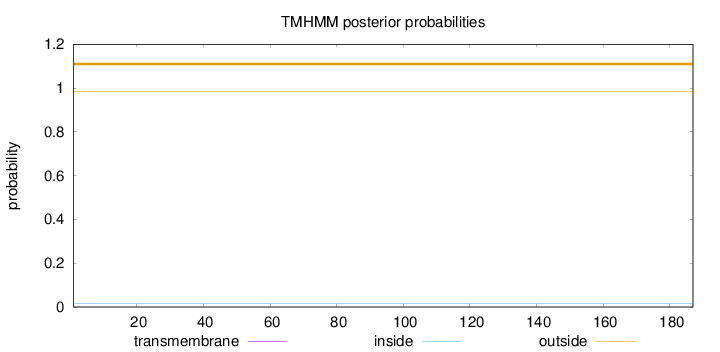
Length:
187
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00023
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01624
outside
1 - 187
Population Genetic Test Statistics
Pi
13.02076
Theta
16.696163
Tajima's D
-1.093427
CLR
35.415711
CSRT
0.121993900304985
Interpretation
Uncertain
