Gene
KWMTBOMO11096 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008295
Annotation
vacuolar_ATP_synthase_catalytic_subunit_A_[Bombyx_mori]
Full name
V-type proton ATPase catalytic subunit A
+ More
V-type proton ATPase catalytic subunit A isoform 2
V-type proton ATPase catalytic subunit A isoform 2
Alternative Name
V-ATPase 69 kDa subunit
Vacuolar proton pump subunit alpha
V-ATPase 69 kDa subunit 2
Vacuolar proton pump subunit alpha 2
Vacuolar proton pump subunit alpha
V-ATPase 69 kDa subunit 2
Vacuolar proton pump subunit alpha 2
Location in the cell
Cytoplasmic Reliability : 2.07
Sequence
CDS
ATGGCGAGCAAAGGCGGTTTGAGGACGATCGCCAATGAGGAGAATGAGGAGAGGTTCGGATATGTCTTCGCCGTATCTGGACCCGTCGTCACTGCCGAAAAGATGTCCGGATCTGCTATGTACGAGTTGGTCCGTGTCGGTTACAATGAACTCGTCGGTGAGATTATCCGTCTTGAAGGTGACATGGCCACCATCCAGGTATACGAAGAAACTTCAGGTGTAACTGTAGGTGATCCAGTACTCCGTACTGGAAAGCCTTTGTCAGTAGAACTTGGTCCCGGTATCTTGGGTTCCATCTTTGACGGTATTCAGCGTCCCCTCAAGGACATCAACGAGCTGACTCAGTCCATCTACATCCCCAAGGGTATCAACGTGCCTTCCCTGGCCAGGGAGGTTGACTGGGAATTTAACCCATTAAATGTTAAGGTCGGGTCCCACATCACCGGTGGAGATTTGTATGGTATTGTACACGAGAACACTCTGGTCAAGCACAGGATGTTGGTCCCGCCCAAAGCCAAGGGAACAGTTACCTATATCGCACCGGCCGGGAACTACAAAGTCACAGACGTGGTGCTGGAGACGGAGTTCGACGGTGAGCGGCAGAAGTACAGCATGTTGCAGGTGTGGCCCGTGCGCCAGCCGCGCCCCGTCACCGAGAAGCTGCCCGCCAACCACCCGCTGCTCACCGGGCAGCGCGTGCTCGACTCGCTCTTCCCTTGCGTGCAGGGAGGTACCACGGCCATCCCCGGGGCTTTCGGTTGCGGCAAAACTGTCATCTCCCAAGCCCTGTCCAAGTACTCCAACTCTGACGTCATCATCTACGTCGGCTGCGGCGAGCGCGGTAACGAGATGTCCGAGGTGCTCCGCGACTTCCCGGAGCTGACGGTGGAGATCGAGGGCGTGACGGAGTCCATCATGAAGCGCACGGCGCTGGTGGCCAACACCTCCAACATGCCCGTGGCGGCGCGCGAGGCTTCCATCTATACCGGCATCACGCTGTCGGAGTACTTCCGCGACATGGGCTACAACGTGTCGATGATGGCGGACTCGACGTCCCGTTGGGCGGAGGCGCTGCGTGAGATCTCTGGTCGCCTGGCGGAGATGCCGGCCGACTCGGGCTACCCCGCCTACCTGGGCGCGCGCCTCGCCTCCTTCTACGAGCGCGCCGGCAGGGTCAAGTGCCTCGGCAACCCGGACCGGGAAGGCTCGGTGTCCATCGTGGGCGCGGTGTCGCCGCCCGGCGGAGACTTCTCGGACCCGGTGACGGCGGCCACGCTCGGCATCGTGCAGGTGTTCTGGGGGCTCGACAAGAAGCTGGCGCAGCGGAAGCACTTCCCCTCCATCAACTGGCTCATTTCATACAGCAAGTACATGCGCGCCCTCGACGACTTCTACGAGAAGAACTACCCCGAGTTCGTCCCGCTCAGGACCAAGGTCAAGGAGATCCTGCAGGAGGAAGAGGACCTGTCCGAAATCGTGCAGCTGGTCGGCAAGGCGTCCCTCGCGGAAACTGACAAGATCACGCTCGAGGTCGCCAAGCTGCTCAAAGACGACTTCCTGCAACAGAACAGCTATTCGTCGTACGATCGCTTCTGCCCGTTCTACAAGACGGTGGGCATGCTGAAGAACATCATCACGTTCTACGACATGTCGCGGCACGCGGTGGAGTCCACGGCGCAGTCCGACAACAAGGTCACGTGGAACGTCATCCGCGACGCCATGGGCAACGTGCTCTACCAGCTCTCCTCCATGAAGTTCAAGGACCCAGTGAAAGACGGTGAACCTAAGATCAAGGCCGATTTCGACCAGCTTTTAGAAGATATGTCTGCCGCTTTCCGCAACCTCGAGGACTAA
Protein
MASKGGLRTIANEENEERFGYVFAVSGPVVTAEKMSGSAMYELVRVGYNELVGEIIRLEGDMATIQVYEETSGVTVGDPVLRTGKPLSVELGPGILGSIFDGIQRPLKDINELTQSIYIPKGINVPSLAREVDWEFNPLNVKVGSHITGGDLYGIVHENTLVKHRMLVPPKAKGTVTYIAPAGNYKVTDVVLETEFDGERQKYSMLQVWPVRQPRPVTEKLPANHPLLTGQRVLDSLFPCVQGGTTAIPGAFGCGKTVISQALSKYSNSDVIIYVGCGERGNEMSEVLRDFPELTVEIEGVTESIMKRTALVANTSNMPVAAREASIYTGITLSEYFRDMGYNVSMMADSTSRWAEALREISGRLAEMPADSGYPAYLGARLASFYERAGRVKCLGNPDREGSVSIVGAVSPPGGDFSDPVTAATLGIVQVFWGLDKKLAQRKHFPSINWLISYSKYMRALDDFYEKNYPEFVPLRTKVKEILQEEEDLSEIVQLVGKASLAETDKITLEVAKLLKDDFLQQNSYSSYDRFCPFYKTVGMLKNIITFYDMSRHAVESTAQSDNKVTWNVIRDAMGNVLYQLSSMKFKDPVKDGEPKIKADFDQLLEDMSAAFRNLED
Summary
Description
Catalytic subunit of the peripheral V1 complex of vacuolar ATPase. V-ATPase vacuolar ATPase is responsible for acidifying a variety of intracellular compartments in eukaryotic cells.
Catalytic subunit of the peripheral V1 complex of vacuolar ATPase. V-ATPase vacuolar ATPase is responsible for acidifying a variety of intracellular compartments in eukaryotic cells (By similarity).
Catalytic subunit of the peripheral V1 complex of vacuolar ATPase. V-ATPase vacuolar ATPase is responsible for acidifying a variety of intracellular compartments in eukaryotic cells (By similarity).
Catalytic Activity
ATP + 4 H(+)(in) + H2O = ADP + 5 H(+)(out) + phosphate
Subunit
V-ATPase is a heteromultimeric enzyme composed of a peripheral catalytic V1 complex (main components: subunits A, B, C, D, E, and F) attached to an integral membrane V0 proton pore complex (main component: the proteolipid protein).
Similarity
Belongs to the ATPase alpha/beta chains family.
Keywords
ATP-binding
Hydrogen ion transport
Ion transport
Nucleotide-binding
Translocase
Transport
Complete proteome
Reference proteome
Phosphoprotein
Feature
chain V-type proton ATPase catalytic subunit A
Uniprot
A1E9B3
P31400
A0A194QGV7
A0A345BEK9
A0A3S2LHJ5
E3W6T9
+ More
A0A212FKI1 A0A221SMT9 A0A2A4J831 A0A0D3QSI4 A0A1S5XVQ3 A0A2H1V5J4 A0A194RIZ0 A0A1L1YM05 A0A0R6XDG1 A0A0J7NF99 E9IUV8 A0A154PQS0 A0A195B5T9 A0A158NIQ0 A0A151XBN6 A0A0L7QN21 A0A195ETN1 E2C1G9 F4WCI9 E2A6I4 A0A3G1SUZ4 A0A0C9Q707 A0A195ELY4 K7INR1 U5EZF7 A0A084VRH3 A0A1B0EXF9 A0A195C3T4 A0A026VXV0 A0A232EWD6 A0A182J067 A0A182NSF3 A0A182QWW0 A0A182XVI8 A0A182W2R7 A0A2M3ZZX2 A0A182FQV4 A0A182MTK0 A0A182RYA8 B0WKP1 W5J9Z0 A0A2M3YYI5 B4KGL4 A0A1L8DF99 A0A1Q3FIB4 A0A088ANZ0 A0A182U8P0 A0A182LC98 A0A182XE81 F5HL05 A0A182HJ42 Q5TTG1 A0A182UMF4 A0A2A3ESC8 V9IIW4 A0A1Q3FIA2 A0A0M3QTT6 B3MNH8 B4IED4 B3N3B5 A4V0N4 Q27331 A0A1W4W9E2 B4MWL4 A0A2Z6ZMA6 B4P2K3 A0A2J7RCG3 Q2TJ56 O16109 A0A0N0BFS9 A0A0K8TQ05 B4JA39 A0A1W4X1V7 B4LSZ1 A0A1B6IZ54 W8B182 A0A3B0JI49 A0A0A1XRY6 A0A1S3CY29 A0A034W5M8 A0A067RPA9 A0A1A9VY13 A0A1A9ZX68 D3TLI6 A0A1A9YB27 A0A1B0BHL1 A0A1A9WYJ2 D2A198 T1PBZ7 B5DJE5 A0A0L0CC76 A0A1B6DHJ3 T1DE30 A0A0K8TWS8
A0A212FKI1 A0A221SMT9 A0A2A4J831 A0A0D3QSI4 A0A1S5XVQ3 A0A2H1V5J4 A0A194RIZ0 A0A1L1YM05 A0A0R6XDG1 A0A0J7NF99 E9IUV8 A0A154PQS0 A0A195B5T9 A0A158NIQ0 A0A151XBN6 A0A0L7QN21 A0A195ETN1 E2C1G9 F4WCI9 E2A6I4 A0A3G1SUZ4 A0A0C9Q707 A0A195ELY4 K7INR1 U5EZF7 A0A084VRH3 A0A1B0EXF9 A0A195C3T4 A0A026VXV0 A0A232EWD6 A0A182J067 A0A182NSF3 A0A182QWW0 A0A182XVI8 A0A182W2R7 A0A2M3ZZX2 A0A182FQV4 A0A182MTK0 A0A182RYA8 B0WKP1 W5J9Z0 A0A2M3YYI5 B4KGL4 A0A1L8DF99 A0A1Q3FIB4 A0A088ANZ0 A0A182U8P0 A0A182LC98 A0A182XE81 F5HL05 A0A182HJ42 Q5TTG1 A0A182UMF4 A0A2A3ESC8 V9IIW4 A0A1Q3FIA2 A0A0M3QTT6 B3MNH8 B4IED4 B3N3B5 A4V0N4 Q27331 A0A1W4W9E2 B4MWL4 A0A2Z6ZMA6 B4P2K3 A0A2J7RCG3 Q2TJ56 O16109 A0A0N0BFS9 A0A0K8TQ05 B4JA39 A0A1W4X1V7 B4LSZ1 A0A1B6IZ54 W8B182 A0A3B0JI49 A0A0A1XRY6 A0A1S3CY29 A0A034W5M8 A0A067RPA9 A0A1A9VY13 A0A1A9ZX68 D3TLI6 A0A1A9YB27 A0A1B0BHL1 A0A1A9WYJ2 D2A198 T1PBZ7 B5DJE5 A0A0L0CC76 A0A1B6DHJ3 T1DE30 A0A0K8TWS8
EC Number
7.1.2.2
Pubmed
1532941
26354079
22118469
28067269
21282665
21347285
+ More
20798317 21719571 20075255 24438588 24508170 30249741 28648823 25244985 20920257 23761445 17994087 20966253 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 9050231 12537569 18327897 26483478 17550304 16579815 9397516 17510324 26369729 24495485 25830018 25348373 24845553 20353571 18362917 19820115 25315136 15632085 26108605 24330624
20798317 21719571 20075255 24438588 24508170 30249741 28648823 25244985 20920257 23761445 17994087 20966253 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 9050231 12537569 18327897 26483478 17550304 16579815 9397516 17510324 26369729 24495485 25830018 25348373 24845553 20353571 18362917 19820115 25315136 15632085 26108605 24330624
EMBL
EF114391
ABL61508.1
X64233
KQ458880
KPJ04654.1
MF370364
+ More
AXF48683.1 RSAL01000118 RVE46789.1 HQ434762 HQ540514 FR727328 ADP23923.1 ADT80587.1 CBY05457.1 AGBW02008030 OWR54253.1 KX668536 ASN77692.1 NWSH01002462 PCG68245.1 KP090287 AJQ81220.1 KX685519 AQQ72785.1 ODYU01000763 SOQ36046.1 KQ460154 KPJ17305.1 KM434187 AKM20771.1 KP341655 AKK21114.1 LBMM01005788 KMQ91205.1 GL766058 EFZ15643.1 KQ435007 KZC13470.1 KQ976582 KYM79851.1 ADTU01001828 KQ982327 KYQ57708.1 KQ414860 KOC60052.1 KQ981979 KYN31267.1 GL451937 EFN78145.1 GL888070 EGI68169.1 GL437132 EFN70941.1 MG243596 AXU38341.1 GBYB01002703 GBYB01009918 JAG72470.1 JAG79685.1 KQ978739 KYN28889.1 GANO01000879 JAB58992.1 ATLV01015669 KE525026 KFB40567.1 AJWK01033992 KQ978292 KYM95517.1 KK107796 QOIP01000013 EZA47674.1 RLU15346.1 NNAY01001876 OXU22668.1 AXCN02000237 GGFK01000734 MBW34055.1 AXCM01000024 DS231974 EDS29972.1 ADMH02001863 ETN60811.1 GGFM01000537 MBW21288.1 CH933807 EDW13214.1 GFDF01008976 JAV05108.1 GFDL01007745 JAV27300.1 AAAB01008879 EGK96966.1 EGK96967.1 EGK96968.1 APCN01002124 EAA44781.1 KZ288193 PBC34029.1 JR049572 AEY61005.1 GFDL01007734 JAV27311.1 CP012523 ALC39449.1 CH902620 EDV32086.1 CH480831 EDW45961.1 CH954177 EDV58755.1 AE014134 BT124897 AAF53232.1 AAF53233.1 ADG57804.1 AFH03689.1 AGB92959.1 AGB92960.1 U59146 U59147 AY084150 CH963857 EDW76155.1 KQ560276 KXJ83361.1 CM000157 EDW88233.1 NEVH01005885 PNF38520.1 AY864912 AF008922 CH477534 KQ435796 KOX73541.1 GDAI01001390 JAI16213.1 CH916368 EDW03713.1 CH940649 EDW63822.1 GECU01015503 JAS92203.1 GAMC01014303 GAMC01014302 JAB92252.1 OUUW01000006 SPP82084.1 GBXI01008949 GBXI01000213 JAD05343.1 JAD14079.1 GAKP01009360 GAKP01009358 JAC49592.1 KK852506 KDR22470.1 CCAG010015982 EZ422288 ADD18564.1 JXJN01014423 KQ971338 EFA02908.1 KA646209 AFP60838.1 CH379061 EDY70417.1 JRES01000622 KNC29832.1 GEDC01012176 JAS25122.1 GALA01001222 JAA93630.1 GDHF01033769 GDHF01020925 GDHF01003060 JAI18545.1 JAI31389.1 JAI49254.1
AXF48683.1 RSAL01000118 RVE46789.1 HQ434762 HQ540514 FR727328 ADP23923.1 ADT80587.1 CBY05457.1 AGBW02008030 OWR54253.1 KX668536 ASN77692.1 NWSH01002462 PCG68245.1 KP090287 AJQ81220.1 KX685519 AQQ72785.1 ODYU01000763 SOQ36046.1 KQ460154 KPJ17305.1 KM434187 AKM20771.1 KP341655 AKK21114.1 LBMM01005788 KMQ91205.1 GL766058 EFZ15643.1 KQ435007 KZC13470.1 KQ976582 KYM79851.1 ADTU01001828 KQ982327 KYQ57708.1 KQ414860 KOC60052.1 KQ981979 KYN31267.1 GL451937 EFN78145.1 GL888070 EGI68169.1 GL437132 EFN70941.1 MG243596 AXU38341.1 GBYB01002703 GBYB01009918 JAG72470.1 JAG79685.1 KQ978739 KYN28889.1 GANO01000879 JAB58992.1 ATLV01015669 KE525026 KFB40567.1 AJWK01033992 KQ978292 KYM95517.1 KK107796 QOIP01000013 EZA47674.1 RLU15346.1 NNAY01001876 OXU22668.1 AXCN02000237 GGFK01000734 MBW34055.1 AXCM01000024 DS231974 EDS29972.1 ADMH02001863 ETN60811.1 GGFM01000537 MBW21288.1 CH933807 EDW13214.1 GFDF01008976 JAV05108.1 GFDL01007745 JAV27300.1 AAAB01008879 EGK96966.1 EGK96967.1 EGK96968.1 APCN01002124 EAA44781.1 KZ288193 PBC34029.1 JR049572 AEY61005.1 GFDL01007734 JAV27311.1 CP012523 ALC39449.1 CH902620 EDV32086.1 CH480831 EDW45961.1 CH954177 EDV58755.1 AE014134 BT124897 AAF53232.1 AAF53233.1 ADG57804.1 AFH03689.1 AGB92959.1 AGB92960.1 U59146 U59147 AY084150 CH963857 EDW76155.1 KQ560276 KXJ83361.1 CM000157 EDW88233.1 NEVH01005885 PNF38520.1 AY864912 AF008922 CH477534 KQ435796 KOX73541.1 GDAI01001390 JAI16213.1 CH916368 EDW03713.1 CH940649 EDW63822.1 GECU01015503 JAS92203.1 GAMC01014303 GAMC01014302 JAB92252.1 OUUW01000006 SPP82084.1 GBXI01008949 GBXI01000213 JAD05343.1 JAD14079.1 GAKP01009360 GAKP01009358 JAC49592.1 KK852506 KDR22470.1 CCAG010015982 EZ422288 ADD18564.1 JXJN01014423 KQ971338 EFA02908.1 KA646209 AFP60838.1 CH379061 EDY70417.1 JRES01000622 KNC29832.1 GEDC01012176 JAS25122.1 GALA01001222 JAA93630.1 GDHF01033769 GDHF01020925 GDHF01003060 JAI18545.1 JAI31389.1 JAI49254.1
Proteomes
UP000053268
UP000283053
UP000007151
UP000218220
UP000053240
UP000036403
+ More
UP000076502 UP000078540 UP000005205 UP000075809 UP000053825 UP000078541 UP000008237 UP000007755 UP000000311 UP000078492 UP000002358 UP000030765 UP000092461 UP000078542 UP000053097 UP000279307 UP000215335 UP000075880 UP000075884 UP000075886 UP000076408 UP000075920 UP000069272 UP000075883 UP000075900 UP000002320 UP000000673 UP000009192 UP000005203 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000075903 UP000242457 UP000092553 UP000007801 UP000001292 UP000008711 UP000000803 UP000192221 UP000007798 UP000249989 UP000002282 UP000235965 UP000069940 UP000008820 UP000053105 UP000001070 UP000192223 UP000008792 UP000268350 UP000079169 UP000027135 UP000078200 UP000092445 UP000092444 UP000092443 UP000092460 UP000091820 UP000007266 UP000095301 UP000001819 UP000037069
UP000076502 UP000078540 UP000005205 UP000075809 UP000053825 UP000078541 UP000008237 UP000007755 UP000000311 UP000078492 UP000002358 UP000030765 UP000092461 UP000078542 UP000053097 UP000279307 UP000215335 UP000075880 UP000075884 UP000075886 UP000076408 UP000075920 UP000069272 UP000075883 UP000075900 UP000002320 UP000000673 UP000009192 UP000005203 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000075903 UP000242457 UP000092553 UP000007801 UP000001292 UP000008711 UP000000803 UP000192221 UP000007798 UP000249989 UP000002282 UP000235965 UP000069940 UP000008820 UP000053105 UP000001070 UP000192223 UP000008792 UP000268350 UP000079169 UP000027135 UP000078200 UP000092445 UP000092444 UP000092443 UP000092460 UP000091820 UP000007266 UP000095301 UP000001819 UP000037069
Interpro
IPR024034
ATPase_F1/V1_b/a_C
+ More
IPR036121 ATPase_F1/V1/A1_a/bsu_N_sf
IPR023366 ATP_synth_asu-like_sf
IPR031686 ATP-synth_a_Xtn
IPR004100 ATPase_F1/V1/A1_a/bsu_N
IPR005725 ATPase_V1-cplx_asu
IPR027417 P-loop_NTPase
IPR020003 ATPase_a/bsu_AS
IPR000194 ATPase_F1/V1/A1_a/bsu_nucl-bd
IPR022878 V-ATPase_asu
IPR031962 DUF4781
IPR036121 ATPase_F1/V1/A1_a/bsu_N_sf
IPR023366 ATP_synth_asu-like_sf
IPR031686 ATP-synth_a_Xtn
IPR004100 ATPase_F1/V1/A1_a/bsu_N
IPR005725 ATPase_V1-cplx_asu
IPR027417 P-loop_NTPase
IPR020003 ATPase_a/bsu_AS
IPR000194 ATPase_F1/V1/A1_a/bsu_nucl-bd
IPR022878 V-ATPase_asu
IPR031962 DUF4781
Gene 3D
ProteinModelPortal
A1E9B3
P31400
A0A194QGV7
A0A345BEK9
A0A3S2LHJ5
E3W6T9
+ More
A0A212FKI1 A0A221SMT9 A0A2A4J831 A0A0D3QSI4 A0A1S5XVQ3 A0A2H1V5J4 A0A194RIZ0 A0A1L1YM05 A0A0R6XDG1 A0A0J7NF99 E9IUV8 A0A154PQS0 A0A195B5T9 A0A158NIQ0 A0A151XBN6 A0A0L7QN21 A0A195ETN1 E2C1G9 F4WCI9 E2A6I4 A0A3G1SUZ4 A0A0C9Q707 A0A195ELY4 K7INR1 U5EZF7 A0A084VRH3 A0A1B0EXF9 A0A195C3T4 A0A026VXV0 A0A232EWD6 A0A182J067 A0A182NSF3 A0A182QWW0 A0A182XVI8 A0A182W2R7 A0A2M3ZZX2 A0A182FQV4 A0A182MTK0 A0A182RYA8 B0WKP1 W5J9Z0 A0A2M3YYI5 B4KGL4 A0A1L8DF99 A0A1Q3FIB4 A0A088ANZ0 A0A182U8P0 A0A182LC98 A0A182XE81 F5HL05 A0A182HJ42 Q5TTG1 A0A182UMF4 A0A2A3ESC8 V9IIW4 A0A1Q3FIA2 A0A0M3QTT6 B3MNH8 B4IED4 B3N3B5 A4V0N4 Q27331 A0A1W4W9E2 B4MWL4 A0A2Z6ZMA6 B4P2K3 A0A2J7RCG3 Q2TJ56 O16109 A0A0N0BFS9 A0A0K8TQ05 B4JA39 A0A1W4X1V7 B4LSZ1 A0A1B6IZ54 W8B182 A0A3B0JI49 A0A0A1XRY6 A0A1S3CY29 A0A034W5M8 A0A067RPA9 A0A1A9VY13 A0A1A9ZX68 D3TLI6 A0A1A9YB27 A0A1B0BHL1 A0A1A9WYJ2 D2A198 T1PBZ7 B5DJE5 A0A0L0CC76 A0A1B6DHJ3 T1DE30 A0A0K8TWS8
A0A212FKI1 A0A221SMT9 A0A2A4J831 A0A0D3QSI4 A0A1S5XVQ3 A0A2H1V5J4 A0A194RIZ0 A0A1L1YM05 A0A0R6XDG1 A0A0J7NF99 E9IUV8 A0A154PQS0 A0A195B5T9 A0A158NIQ0 A0A151XBN6 A0A0L7QN21 A0A195ETN1 E2C1G9 F4WCI9 E2A6I4 A0A3G1SUZ4 A0A0C9Q707 A0A195ELY4 K7INR1 U5EZF7 A0A084VRH3 A0A1B0EXF9 A0A195C3T4 A0A026VXV0 A0A232EWD6 A0A182J067 A0A182NSF3 A0A182QWW0 A0A182XVI8 A0A182W2R7 A0A2M3ZZX2 A0A182FQV4 A0A182MTK0 A0A182RYA8 B0WKP1 W5J9Z0 A0A2M3YYI5 B4KGL4 A0A1L8DF99 A0A1Q3FIB4 A0A088ANZ0 A0A182U8P0 A0A182LC98 A0A182XE81 F5HL05 A0A182HJ42 Q5TTG1 A0A182UMF4 A0A2A3ESC8 V9IIW4 A0A1Q3FIA2 A0A0M3QTT6 B3MNH8 B4IED4 B3N3B5 A4V0N4 Q27331 A0A1W4W9E2 B4MWL4 A0A2Z6ZMA6 B4P2K3 A0A2J7RCG3 Q2TJ56 O16109 A0A0N0BFS9 A0A0K8TQ05 B4JA39 A0A1W4X1V7 B4LSZ1 A0A1B6IZ54 W8B182 A0A3B0JI49 A0A0A1XRY6 A0A1S3CY29 A0A034W5M8 A0A067RPA9 A0A1A9VY13 A0A1A9ZX68 D3TLI6 A0A1A9YB27 A0A1B0BHL1 A0A1A9WYJ2 D2A198 T1PBZ7 B5DJE5 A0A0L0CC76 A0A1B6DHJ3 T1DE30 A0A0K8TWS8
PDB
6O7X
E-value=0,
Score=2134
Ontologies
PATHWAY
GO
PANTHER
Topology
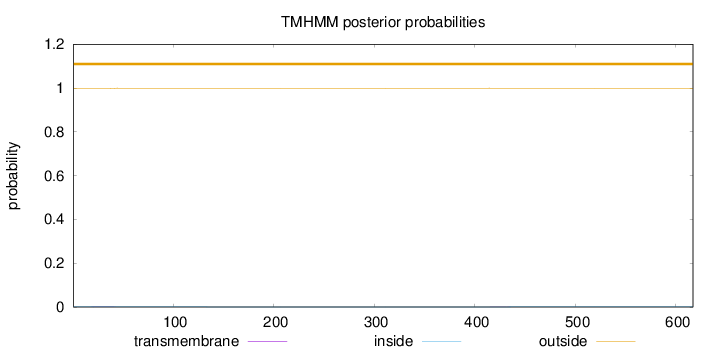
Length:
617
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0532300000000001
Exp number, first 60 AAs:
0.04311
Total prob of N-in:
0.00257
outside
1 - 617
Population Genetic Test Statistics
Pi
209.332352
Theta
172.331838
Tajima's D
0.478363
CLR
10.605234
CSRT
0.512574371281436
Interpretation
Uncertain
